Figure 1. Conservation of viral 2′-O-methyltransferases.
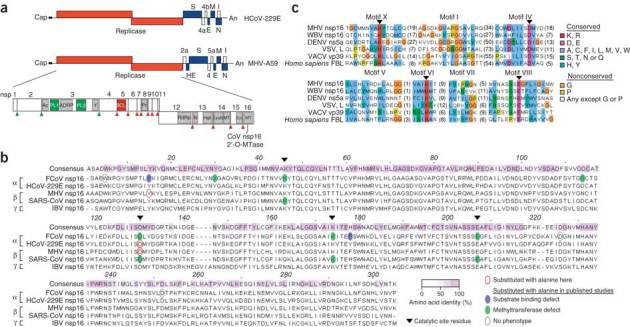
(a) Human and mouse coronavirus genomes, including viral open reading frames (boxes; labels above and below identify gene products). The replicase gene, including conserved domains and viral proteinase cleavage sites (upward arrowheads) that separate nsp1–nsp16, is enlarged. CoV nsp16 MTase, nsp16-associated 2′-O-methyltransferase. (b) ClustalW2 alignment of coronavirus nsp16 amino acid sequences, representative of α-, β- and γ-coronaviruses. Keys (bottom right) identify sequence conservation (amino acid identity), amino acid substitutions and phenotypes of mutant proteins. FCoV, feline coronavirus; MHV, MHV strain A59; SARS-CoV, severe acute respiratory syndrome coronavirus; IBV, infectious bronchitis virus. (c) Comparison of the sequences of viral and cellular methyltransferase motifs. WBV, white bream virus (order Nidovirales); DENV, dengue virus (family Flaviviridae); VSV, vesicular stomatitis virus (order Mononegavirales); VACV, vaccinia virus (family Poxviridae); FBL, fibrillarin (Homo sapiens). Right, key for amino acid similarity (single-letter codes) and conservation according to default ClustalX.
