A fundamental step in linking the activity of individual neurons to circuits and animal behavior is to identify their patterns of connectivity. Traditional methods in electrophysiology and microscopy, although powerful, are limited to addressing only a neuron's immediate neighbors (1). A further limitation of these methods is that this can be addressed only in a piecemeal manner, whereby just a few of a neuron's many synaptic connections can be probed in a given experiment. Determining the larger network connectivity of individual neurons within the intact brain requires transsynaptic tracers. Advances in virology and molecular genetics have improved our ability to target a single neuron and to trace its inputs (2), but what is missing from this toolbox is a reliable method to trace a neuron's output. In a remarkable study in PNAS, Beier et al. present a virus-based anterograde tracer, which targets genetically selected neurons and unambiguously maps the mono- and polysynaptic outputs with relatively low cytotoxicity to the circuit it is tracing (3).
At the beginning of the 20th century, the pioneers in neuroanatomy begun to lay the groundwork for mapping the connections in the central and peripheral nervous system, primarily through the use of fiber degeneration studies carried out in combination with silver impregnation methods. Improvements on this approach were made through the discovery of tracer materials that were directionally transported within the axons (4). Beginning with the finding that the plant enzyme HRP is taken up by the axons and actively transported back to the soma, neuroanatomists have used a variety of tracers ranging from plant lectins and radioactively labeled amino acids to bacterial toxins and fluorescent dyes. These chemical tracers can reliably reveal the locations of neurons projecting to or from particular brain regions, perhaps most memorably through the demonstration of ocular dominance columns in the visual cortex. However, with the exception of wheat germ agglutinin–leptin and tetanus toxin, most chemical tracers do not cross synapses and hence are limited to examining neuronal connections in a point-to-point manner. Furthermore, because of the tracer material's dependence on cellular machinery for uptake and transport, neuroanatomical tracers produce an incomplete diagram of a labeled neuron's afferent and efferent connectivity (5).
The recognition that several viruses entered the central nervous system from the periphery by “hitchhiking” along synaptically connected neurons provided the first clue that neurotrophic viruses would be a valuable alternative to chemical tracers (6). The neurotrophic viruses function as self-amplifying tracers that can produce intense labeling in the infected recipient neurons, distinguishing them from the majority of chemical tracers that are quickly diluted. Several strains of α-herpesviruses, for which the directionality of the viral transport has been shown to depend on the specific strain, are commonly used viruses for tracing neuronal connections (7). In addition, the recognition that the attenuated strains of rabies virus (RV) have a strong affinity for neurons and travels exclusively in the retrograde direction through chemical synapses established them as an attractive retrograde transsynaptic tracer (8). Four years ago, Callaway and coworkers presented a dramatic improvement in our ability to direct the synaptic specificity of the RV-based transsynaptic tracers (9). They demonstrated a clever approach for genetically targeting neuronal subsets for RV infection while also restricting viral propagation to monosynaptic targets through RV glycoprotein (G) complementation. From a conceptual standpoint, this study also supported previous observations that the G coat particle alone is necessary and sufficient to alter the transport properties and tropism of neurotropic viruses. This finding suggested to Cepko and coworkers, who have a long history of using viruses in a creative manner in the nervous system (10), that the virus G primes the virus for retrograde or anterograde transport within the cell as well as entry into connected neurons.
To exploit the potential power of envelope proteins to control the direction of synaptic viral transport, Cepko and coworkers (3) attempted to engineer a viral vector that would be transmitted transsynaptically in defined directions depending on the viral G it encodes. As a vector for viral tracing, the authors turned to vesicular stomatitis virus (VSV). This virus is a negative strand RNA virus that is a member of the Rhabdoviridae family and has properties such as the ability to produce in high titers with relatively low toxicity that make it an attractive target for such applications. VSV does not exhibit directional synaptic transport, which serves as an advantage for VSV as a passive gene delivery vector. They have substituted VSV-G with one of several Gs from other viruses in conjunction with a reporter gene (YFP) to provide an easy method for visualizing synaptically connected neurons (Fig. 1). Comparing the VSV's directionality with different glycoproteins, Beier et al. (3) show that replacing the host G with RV-G endows the VSV with the ability to spread retrogradely, consistent with previous findings (9). Remarkably, the authors observed that, when VSV expresses on its surface the G from lymphocytic choriomeningitis virus (LCMV), an arenavirus, it travels exclusively in the anterograde direction.
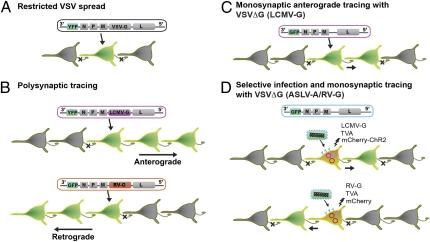
Fig. 1.
Schematic description of the transneuronal tracers generated by Beier et al. (3) in PNAS. (A) The VSV genome was modified such that a fluorescent reporter (YFP) was placed upstream of viral proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), G, and polymerase (L). VSV with endogenous G did not display transsynaptic spread. (B) Directionally selective polysynaptic tracers were generated by replacing the VSV-G with LCMV-G (purple) for anterograde spread, or with RV-G (orange) for retrograde spread of the viral vector. (C) G protein-deficient VSV (VSVΔG) pseudotyped with LCMV-G exhibited monosynaptic anterograde spread. (D) ASLV-A/RV-G pseudotyped VSVΔG can only infect the neurons expressing the TVA receptor from biolistic transfection (blue). By cotransfecting the gene for the LCMV-G in the same neurons, the VSVΔG can produce viruses that were able to transsynaptically infect postsynaptic neurons (Upper). VSVΔG repackaged with the G coat protein expressed from RV-G plasmid (orange) displayed transsynaptic spread to the presynaptic population of neurons (Lower). Because only the initially infected neuron contains virus G protein (LCMV-G or RV-G), VSV cannot spread any further, limiting VSV infection to neurons monosynaptically connected to initially infected population. A plasmid encoding mCherry was cotransfected for the cell originally targeted for infection to be identified within the monosynaptic network of GFP-labeled cells.
As proof of principle, the authors tested the synaptic specificity of this approach by tracing well described circuits in the visual, striatal, and olfactory systems. In vivo inoculation of the replication competent VSV(LCMV-G) into vitreous humor of the eye, caudate putamen, or the olfactory epithelium, all showed transsynaptic labeling of neurons known to receive afferent synaptic connections from neurons in the injection site. In none of the traced circuits—even the shorter-distance connections such as those in the striatum—did the VSV(LCMV-G) exhibit retrograde spread. Moreover, the VSV(LCMV-G) appeared to preferentially label neurons rather than progenitor or glial cells, with the exception of nonspecifically infection of immature neurons in the rostral migratory stream. Additionally, as the VSV virus encoding the LCMV-G is amplified through replication and packaging in the recipient neurons, the anterograde labeling could proceed several synaptic steps away from the primary recipient neurons without dilution in a time-dependent manner (one synapse per day).
Replication-competent VSV(LCMV-G) can trace synaptically connected chains of neurons in topographically organized
Beier et al. present a virus-based anterograde tracer.
sensory systems, but fails to distinguish direct from indirect postsynaptic targets. Therefore, Beier et al. (3) undertook a variation of their approach akin to that previously used by Callaway and coworkers (9) to limit VSV infection to neuronal subsets, while restricting viral propagation to direct monosynaptic postsynaptic targets by LCMV-G complementation. Replication-incompetent VSV (i.e., VSVΔG) was pseudotyped with the avian-specific ASLA-A/RV-G fusion protein, rendering the recombinant virus unable to infect mammalian cells, unless complemented with the receptor for this protein: TVA. To allow for anterograde transport, the TVA construct was presented in conjunction with LCMV-G into cultured hippocampal slices by gene-gun transfection. For visualization and electrophysiological validation of synaptically connected pairs, the authors also transfected the cultured neurons with channelrhodopsin-2, which allowed the initially infected/transfected neurons to be stimulated by light. Indeed, the majority of the downstream neurons exhibited time-locked synaptic currents upon light stimulation of the presynaptic neuron, whereas none of the noninfected neurons in the vicinity were activated upon stimulation of the infected/transfected neuron. These in vitro proof-of-principle experiments provide direct evidence that this approach will permit monosynaptic tracing of the direct outputs of genetically specified cell types, in a manner amenable to physiological study in intact animals. A concern always associated with such approaches is that the very viral vectors that permit tracing of circuits are unfortunately also toxic to the infected populations. In this regard, the authors provide evidence that VSVΔG with a point mutation in the viral matrix protein (M51R) reduces the cytotoxicity of VSV, suggesting that the negative impact of viral toxicity can be minimized if not eliminated. Taken together, there seems no doubt that this approach will be of great use for future in vivo monosynaptic anterograde tracing experiments.
A further observation noted by these investigators (3) is that transsynaptic anterograde spread of replication incompetent VSV pseudotyped with LCMV-G can occur in absence of a functional G in the genome. This likely reflects that the virion particles contain sufficient LCMV-G protein to complement the absence of this gene upon secondary infection of neurons monosynaptically connected to those receiving a primary infection. As a result, this virus might prove valuable in organisms that are not readily accessible to genetic modifications (e.g., primates, songbirds).
The mechanisms responsible for egress and cellular transport of RV or LCMV are not fully understood, and the receptors proposed for directing RV or LCMV infections are too broad for a comprehensive analysis of specific targeting to pre- or postsynaptic sites. However, future developments in the field of virology will ultimately explain the direction specific behaviors of viruses such as the one demonstrated by Cepko and coworkers (3) and continue to enrich the neuroscientists’ toolbox with a fascinating array of virus based circuit tracers.
The stage is now set to tackle one of the most daunting obstacles in the CNS, the examination of neural circuits in living animals. With these methods in hand, it is easy to imagine how the combination of genetic and optogenetic tools can be used to modify or monitor activity in the nervous system (11). Coupled with recombination approaches for providing neuron type specificity (12), circuit based neuroanatomy will herald a new era of investigation.
Footnotes
The authors declare no conflict of interest.
See companion article on page 15414 of issue 37 in volume 108.
References
- 1.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beier KT, et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci USA. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köbbert C, et al. Current concepts in neuroanatomical tracing. Prog Neurobiol. 2000;62:327–351. doi: 10.1016/s0301-0082(00)00019-8. [DOI] [PubMed] [Google Scholar]
- 5.Kuypers HG, Ugolini G. Viruses as transneuronal tracers. Trends Neurosci. 1990;13:71–75. doi: 10.1016/0166-2236(90)90071-h. [DOI] [PubMed] [Google Scholar]
- 6.Enquist LW, Card JP. Recent advances in the use of neurotropic viruses for circuit analysis. Curr Opin Neurobiol. 2003;13:603–606. doi: 10.1016/j.conb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Song CK, Enquist LW, Bartness TJ. New developments in tracing neural circuits with herpesviruses. Virus Res. 2005;111:235–249. doi: 10.1016/j.virusres.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Ugolini G. Advances in viral transneuronal tracing. J Neurosci Methods. 2010;194:2–20. doi: 10.1016/j.jneumeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Wickersham IR, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci USA. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 12.Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci USA. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]