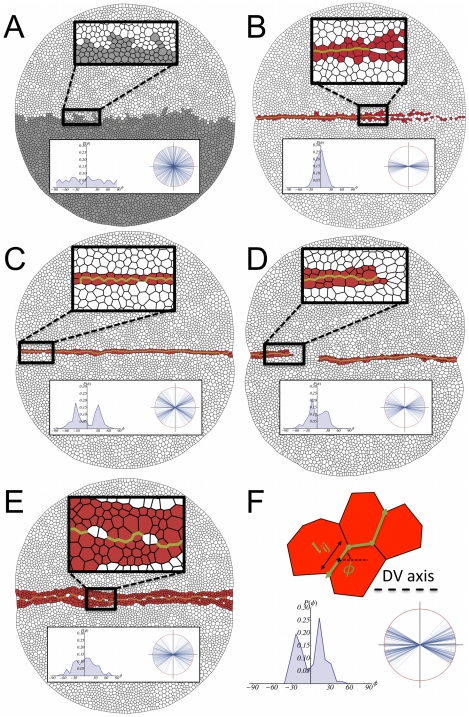
Figure 5. Mutant analysis.
A: In the absence of a DV organizer, cells of opposite compartments intermix. B: The differential cell adhesion is removed in this simulation and as a result the organizer cannot maintain its stability. C: If the actomyosin cable is removed the width of the organizer is reduced at many locations to one cell and is not robust to perturbations. D: A criterion for cell cleavage is required for maintaining the stability of the boundary. If no rule is prescribed (here cells divide at random orientations) then the DV organizer easily breaks leading to cell intermingling between compartments. E: The duration of the cell cycle is set to be the same in the whole disc. This produces a wiggly boundary and a wide organizer. F: In order to quantify the structure of the boundary we evaluate the angle 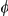 for all cell edges
for all cell edges 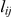 that define the boundary as illustrated on the top of this panel. On the bottom we show the results of the wild-type situation (Fig. 2) in regard to the representation of those angles in a polar plot (right) where all edges lengths are normalized to the unity and their associated histogram (left). The insets in panels A–E reveal that the mutants lead to boundaries/organizers that are weaker and/or wigglier than the wild-type (see text).
that define the boundary as illustrated on the top of this panel. On the bottom we show the results of the wild-type situation (Fig. 2) in regard to the representation of those angles in a polar plot (right) where all edges lengths are normalized to the unity and their associated histogram (left). The insets in panels A–E reveal that the mutants lead to boundaries/organizers that are weaker and/or wigglier than the wild-type (see text).