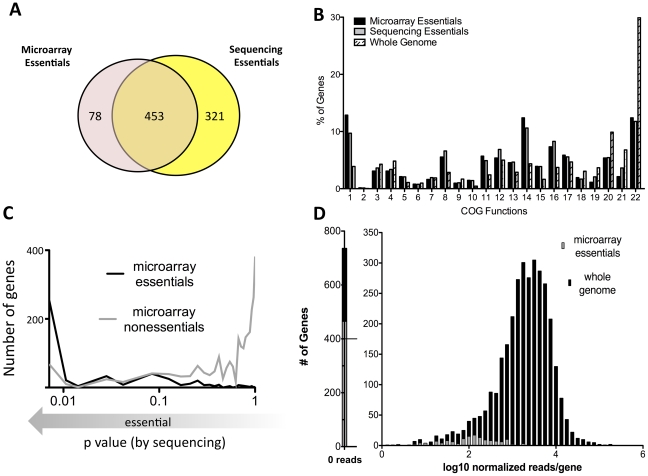
Figure 2. Defining essential genes with deep sequencing is more sensitive and precise than previous approaches.
(A) Gene sets predicted to be essential via microarray [6] are compared with genes predicted to be essential via deep sequencing.(p<0.05) (B) Microarray and deep sequencing identify essential genes sets consisting of similar predicted functions, which differ from the genome as a whole. Clusters of Orthologous Groups (COG) categories associated with essential genes predicted by microarray or deep sequencing (p<0.05) are plotted along with the predictions for the entire genome. COG functions are: 1) Translation, ribosomal structure, biogenesis, 2) RNA processing, modification, 3) Transcription, 4) Replication, recombination, repair, 5) Cell cycle control, cell division, chromosome partitioning, 6) Defense mechanisms, 7) Signal transduction mechanisms, 8) Cell wall/membrane/envelope biogenesis, 9) Cell motility, 10) Intracellular trafficking, secretion, vesicular transport, 11) Posttranslational modification, protein turnover, chaperones, 12) Energy production, conversion, 13) Carbohydrate transport, metabolism, 14) Amino acid transport, metabolism, 15) Nucleotide transport, metabolism, 16) Coenzyme transport, metabolism, 17) Lipid transport, metabolism, 18) Inorganic ion transport, metabolism, 19) Secondary metabolites biosynthesis, transport, catabolism, 20) General function prediction only, 21) Function unknown, 22) No assignment. (C) Microarray predictions are generally, but not absolutely, consistent with deep sequencing. The probability of essentiality defined via deep sequencing is plotted on the x-axis (as log p value) for genes previously predicted by microarray to be either essential (black) or nonessential (gray). (D) Mutations in nonessential genes previously predicted to be required for growth cause quantitative fitness defects. Normalized sequence reads per gene (log scale) are plotted for all genes (black) and genes predicted to be essential by microarray (gray). Previously defined essential genes with detectable insertions are associated with low sequence read counts, suggesting quantitative underrepresentation in the library. Genes containing fewer than seven TA's were excluded from all analyses, since statistically-significant predictions could not be made.