Figure 5.
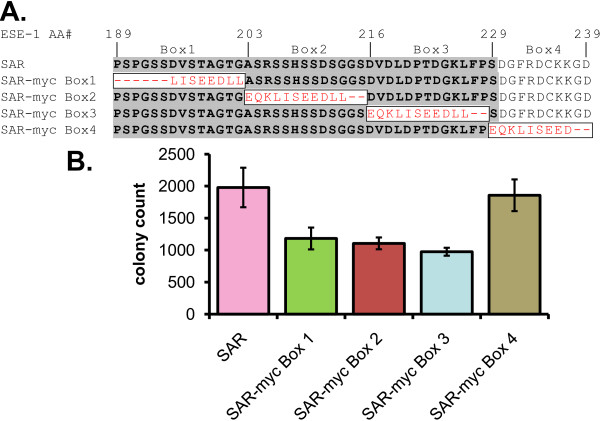
An intact SAR domain is required for optimal transforming activity. (A) Amino acid sequence of the SAR portion of GFP-SAR (SAR, top row) followed by sequences of GFP-SAR-myc Box 1 - Box 4 mutants, containing in-frame replacement of SAR domain amino acids 1-14 (SAR-myc Box 1, replaced AAs correspond to ESE-1 AAs 189-202), SAR amino acids 15-27 (SAR-myc Box 2, replaced AAs correspond to ESE-1 AAs 203-215), SAR amino acids 28-40 (SAR-myc Box 3, replaced AAs correspond to ESE-1 AAs 216-228), and amino acids 41-50, just distal to the SAR domain (SAR-myc Box 4, replaced AAs correspond to ESE-1 AAs 229-239). Black capital letters on gray background - SAR sequence. Black capital letters on white background - carboxy-terminal ESE-1 flanking sequence. Boxed red letters - myc epitope amino acids. Numbers above the SAR sequence indicate corresponding amino acid locations in the ESE-1 protein. (B) Quantification of anchorage-independent colony formation of pooled MCF-12A cell populations stably expressing GFP-SAR-myc Box 1 - Box 4 constructs (SAR-myc Box 1 - Box 4). Three independent, stably transfected MCF-12A populations for each construct were seeded in triplicate soft agarose cultures and colonies visible by 6X bright field microscopy were counted 21 days later. Colony numbers formed by GFP-SAR transfectants (SAR, purple bar), included here to facilitate comparison between SAR-myc Box 1-4 mutants and GFP-SAR control population, are reproduced from Figure 4C. Bars show average numbers of colonies per plate +/- s.e.m.