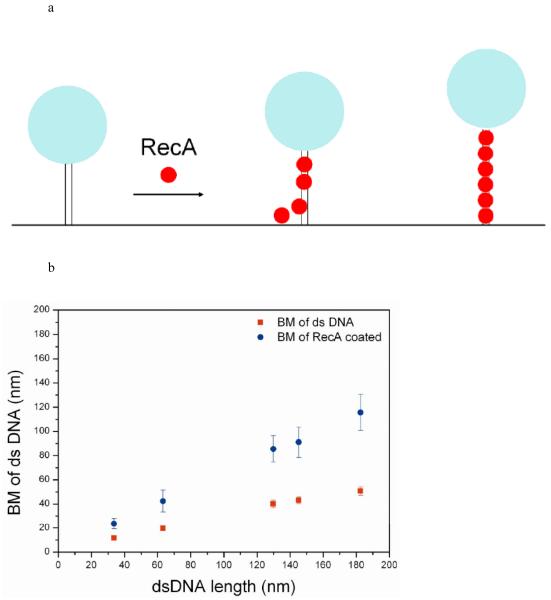
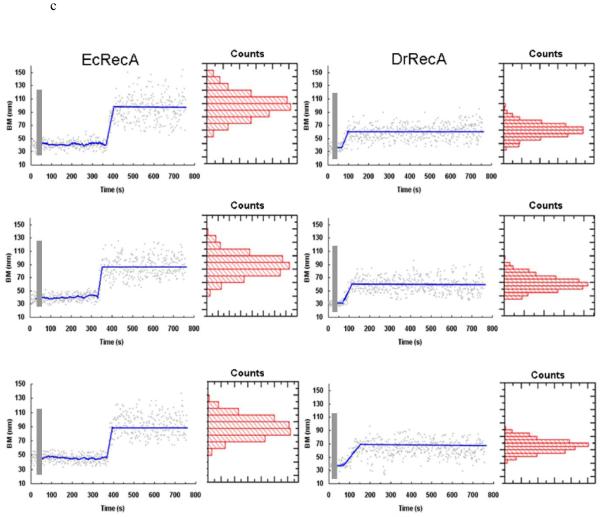
Figure 1.
Observation of RecA assembly along a single duplex DNA molecule using TPM experiments. (a) Changes in bead Brownian motion amplitude reflect the real-time dynamics of RecA nucleation and extension processes. (b) The amplitude of bead’s BM is found to be proportional to bare DNA length (■) within the DNA size studied. The stable extended EcRecA/dsDNA filaments in the presence of the non-hydrolyzable ATP analog, ATPγS (● ), shows a similar linear DNA length dependence, but with a larger slope. Each point contains data from more than 200 tethers and error bars indicate standard deviation. (c) Representative time traces of RecA assembly on a single 382 bp duplex DNA molecule in Buffer B at pH 6.16 and 2 mM ATP. The left column contains traces of EcRecA and the right column for DrRecA. The gray bar designates the time of RecA addition and system re-stabilization (~9-11 s). The recordings were continuous during the buffer change, so the same DNA tethers were monitored throughout the reaction. Time between RecA flow in and Brownian motion change is regarded as nucleation time (the blue line was fit with moving average adjoining 20 points). The phase in which the bead BM showed continuous rise is defined as extension and is fitted with the method described in statistic analysis. We defined the final BM plateau as the maximum BM achieved. BM values of the plateau were collected and the histogram shown beside each time trace. The blue line of the final part indicates the BM of the Gaussian peak.