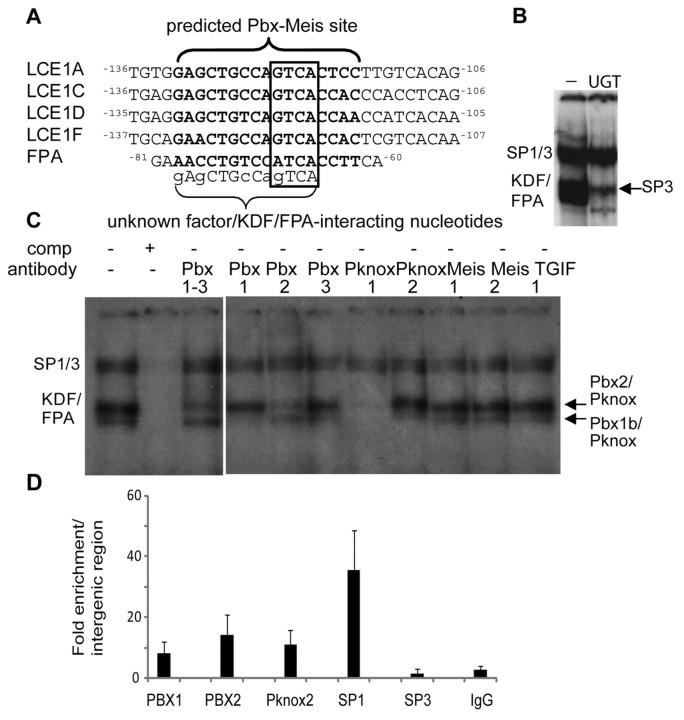
Fig. 3.
TALE proteins PBX1b, PBX2 and Pknox bind the LCE KDF/FPA site. (A) Alignment of the KDF/FPA binding sequences from LCE genes and the involucrin promoter with a PBX–Meis1 experimentally derived matrix consensus binding sequence shown in bold (from MatInspector). PBX–Meis1 core sequences are boxed. Unknown factor(s) or KDF sites from LCE1B, LCE1E and involucrin KDF itself did not meet the stringency criterion for core (LCE1B) or matrix (LCE1E, KDF) similarity. (B) EMSA with the LCE1C promoter fragment and a well characterised PBX site from the UDP glucuronosyltransferase (UGT2B17) gene (Gregory and Mackenzie, 2002) (denoted ‘UGT’; −, no competitor) very effectively competes with KDF/FPA. Competition reveals binding by the truncated SP3 transcription factor. (C) EMSA with TALE-family-specific antibody supershift confirms that TALE family members bind the KDF/FPA site. The two KDF/FPA binding complexes are identified as upper PBX2–Pknox factor and the lower as PBX1b–Pknox factor. (D) ChIP shows that PBX1, PBX2, PKNOX2 and SP1 bind to the LCE1C site in vivo. SP3 binding was not detected. Data is presented as fold enrichment of LCE1C binding (as % of input genomic DNA) compared with enrichment of binding to an intergenic region.