Abstract
Directed evolution and protein engineering approaches used to generate novel or enhanced biomolecular function often use the evolutionary sequence diversity of protein homologs to rationally guide library design. To fully capture this sequence diversity, however, libraries containing millions of variants are often necessary. Screening libraries of this size is often undesirable due to inaccuracies of high-throughput assays, costs and time constraints. The ability to effectively cull sequence diversity while still generating the functional diversity within a library thus holds considerable value. This is particularly relevant when high-throughput assays are not amenable to select/screen for certain biomolecular properties. Here, we summarize our recent attempts to develop an evolution-guided approach, Reconstructing Evolutionary Adaptive Paths (REAP), for directed evolution and protein engineering that exploits phylogenetic and sequence analyses to identify amino acid substitutions that are likely to alter or enhance function of a protein. To demonstrate the utility of this technique, we highlight our previous work with DNA polymerases in which a REAP-designed small library was used to identify a DNA polymerase capable of accepting non-standard nucleosides. We anticipate that the REAP approach will be used in the future to facilitate the engineering of biopolymers with expanded functions and will thus have a significant impact on the developing field of `evolutionary synthetic biology'.
Keywords: directed evolution, evolutionary models, functional divergence, protein engineering
Introduction
Protein engineering and directed evolution are powerful techniques for improving or modifying the activity, specificity and/or stability of proteins (Arnold and Georgiou 2003; Brakmann 2001; Crameri et al. 1998; Lutz and Patrick 2004; Ness et al. 2002). These approaches have been applied to a wide range of protein families for uses in technology development, therapeutics, agriculture and chemistry. The general technique consists of fundamental steps that are repeated until a desired property emerges: 1) the introduction of sequence diversity to produce a library of variants from a parent protein; 2) a screen or selection that identifies the variant(s) with the desired phenotype; and, if necessary, 3) recombination between selected variants to produce new sequence combinations.
The success of these experiments depends on both the sequence/functional diversity sampled and the screening/selection assay. Researchers often design large libraries in order to capture as much functional diversity as possible. However, use of such large libraries requires high-throughput assays to select or screen for the desired functional variants. Ideally, these high-throughput assays efficiently capture protein variants with a preferred biomolecular function. In practice, however, high-throughput assays often capture variants whose behavior only serves as a proxy of a desired function (Ness et al. 2005). The importance of the assay's specificity for measuring a desired quality is evident from the field's axiom of `you get what you select for' (You and Arnold 1996). Therefore, more accurate low-throughput assays would be greatly preferred if library sizes could be reduced without sacrificing functional diversity.
Due to the necessity of low-throughput screening techniques to capture certain protein functions, many research groups have recently focused their attention on library quality instead of quantity in directed evolution and protein-engineering experiments (Lehman and Unrau 2005; Liao et al. 2007; Lutz and Patrick 2004). Smaller pools of variants consisting of fewer and more-focused substitutions are displacing large libraries built from random or shuffled substitutions. The success of these small libraries ultimately depends on their ability to generate a sufficient amount of functional diversity within their reduced sequence space.
Here, we present an approach, termed `Reconstructing Evolutionary Adaptive Paths' (REAP), that uses the evolutionary history of a protein and the functional diversity of extant homologs to guide the design of small libraries that capture meaningful sequence diversity. Previous work with molecular evolutionary models led us to conclude that understanding the evolutionary history of gene families can offer insight into the particular residues of a gene likely to alter function when working with reduced sequence space for small libraries (Gaucher et al. 2002; Gaucher et al. 2001; Gaucher et al. 2003). This strategy can be used to identify residue substitutions that are likely to affect a particular functional property. Thus, a library targeting only these substitutions can be designed to capture functional diversity in a relatively small amount of sequence space.
The REAP approach is distinct from previous methods used to design libraries in that it is more explicit in its use of evolutionary information. Our approach relies on phylogenetic analysis of homologous sequences to detect signatures of functional divergence, and reconstruction of the individual mutations that occurred along these functionally divergent branches of the phylogeny. Here, we present the underlying principles of the REAP approach, an illustration of REAP compared to other common approaches, and demonstrate the power of the REAP approach by presenting a case where it was used to successfully engineer a DNA polymerase capable of utilizing non-standard nucleosides.
Theory
Signatures of functional divergence
Signatures of functional divergence and adaptive evolution can be identified using multiple models of molecular sequence evolution. For instance, we and others have developed a methodology that models site-specific rate shifts under a heterotachous framework (where mutation rates for a given residue are not necessarily constant across a phylogeny) such that the homotachous model (where the site-specific mutation rate remains constant across the phylogeny) can be treated as a special case (Gaucher et al. 2002; Gu 2001; Gu and Vander Velden 2002; Knudsen and Miyamoto 2001; Lopez et al. 2002; Pupko and Galtier 2002; Wang et al. 2007). This provides an opportunity to statistically determine which of the two models better fit the data. Consider a phylogeny with at least two monophyletic clusters, generated by gene duplication or speciation event. It is proposed that a site has two states. In one state (S0), a site has the same mutation rate in both monophyletic clusters; in the other state (S1), a site has different rates between the two clusters. The prediction of functional divergence (θ) between two clusters is defined as the probability of a site being S1, [i.e, θ = P(S1)], which is called the coefficient of evolutionary functional divergence (Gu 2001). With this approach, the homotachous model is a special case when θ = 0. Conceptually, θ measures the degree of independence (i.e., lack of correlation) between the relative evolutionary rates at the sites in one protein subfamily/lineage versus those in another.
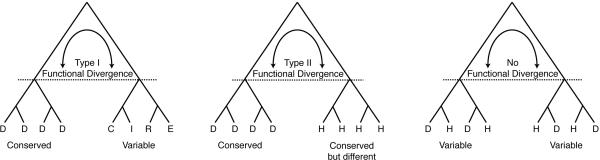
Two types of sequence change are associated with site-specific rate shifts. Both are based on the assumption that residues critical for function tend to be conserved over the course of evolution (Figure 1). The first type of sequence change, type-I functional divergence or heterotachy (also called covarion-like), involves a shift in the relative rate of evolution at a particular site: a site shifts from being relatively strongly conserved (functionally important) to being less conserved (functionally less important) or vice versa. Alternatively, in type-II functional divergence, residues involved in the shift in function are highly conserved in both subfamilies but differ in the identity of their amino acids between the monophyletic subfamilies/lineages. Thus, analyses that combine measurements of θ with amino acid identities at sites across a protein can identify those sites associated with type I and type II functional divergence.
Figure 1.
Signatures of evolutionary functional divergence. Schematics of site-specific types I & II functional divergence between subfamilies of sequences. Left, type-I functional divergence in which a specific site is occupied by a conserved aspartate residue (D) in one lineage but occupied by many residues at the homologous site in the other lineage. Middle, type-II functional divergence in which a specific site is again occupied by a conserved aspartate residue (D) in one lineage while the homologous site in the other lineage is also conserved but occupied by a different residue (histidine, H). Right, no functional divergence associated with the replacements of aspartates and histidines. Functional inferences associated with these patterns require phylogenetic analysis otherwise they are indistinguishable from historical contingence (i.e., common ancestry).
Additional models can also be exploited to identify episodes of adaptive evolution across a phylogeny, thereby identifying sites important for a variant library. In particular, the nonsynonymous-to-synonymous ratio is notable for detecting positive selection although the method can also identify sites predicted to be neutrally evolving (Benner and Gaucher 2001; Bielawski and Yang 2004; Gaucher et al. 2003; Wong et al. 2004). For sequences evolving under a neutral model of evolution, a comparison of the sequences will yield a nonsynonymous/synonymous ratio of ~1. Sequences under purifying/stabilizing selection will display a ratio less than 1, while sequences under diversifying/positive selection display a ratio greater than 1. Recent statistical advances now allow for the identification of either whole genes or specific sites within genes that have undergone positive selection during their evolutionary histories (Bielawski and Yang 2004). Evolutionary analysis of nonsynonymous to synonymous ratios is therefore another powerful statistical technique to identify functionally important residues.
Once molecular evolutionary models have identified sites implicated in functional divergence, one must next identify the individual mutations that occurred during adaptive episodes at these sites through evolutionary history. This can be accomplished using ancestral sequence reconstruction (Gaucher et al. 2008; Yang et al. 1995). The most common approach for ancestral sequence reconstruction utilizes a model-based likelihood (Thornton 2004). The method follows standard Bayesian statistical theory: given the data at a site, the conditional probabilities of different ancestral states can be compared; and the reconstruction having the highest conditional probability is most often the accepted residue at an ancestral position. Although individual mutations occurring along phylogenetic branches implicated in functional divergence can also be determined by manual inspection, this can be difficult, especially when functional divergence is detected using the nonsynonymous to synonymous ratio. As such, the inference of ancestral character states using explicit models of molecular evolution is preferred. The inferred ancestral character states then serve as the sequence information used to design library variants.
Hypothetical example of REAP and traditional library design methods
Identification of the various types of signatures of functional divergence left in the sequence record and inference of the amino acid replacements at these sites creates a powerful tool for variant library design. During the evolutionary divergence of a gene family, members of each lineage collect three types of mutations: 1) those that are responsible for the functional divergence of different lineages or homologs, 2) those that are due to neutral evolutionary forces along a branch, and 3) deleterious mutations either weeded out by natural selection or randomly fixed. While most library designs sample all three types of mutations when shuffling homologous sequences, the REAP approach specifically attempts to design variants that sample from only the first type of mutation, offering the tremendous advantage of culling out the much larger number of neutral or random mutations observed throughout evolution of a protein family. To illustrate how this affects the library size, sequence space and functional space of a variant library, consider the hypothetical library design of a fluorescent protein family using two popular approaches and the REAP approach.
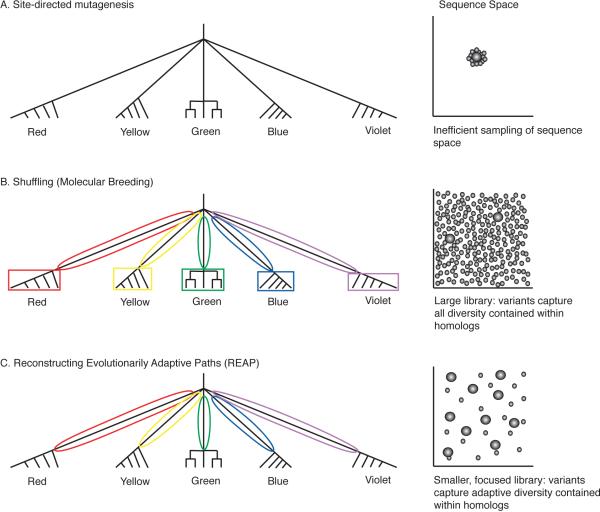
This hypothetical fluorescent protein family contains five homologous subfamilies of individual fluorescent spectra. Each subfamily contains five sequences and all five subfamilies share a common ancestor in the relationship of a polytomy (Figure 2A). The evolution/engineering of fluorescent proteins with novel properties (i.e., a unique emission spectra) can be attempted using libraries designed by methods such as site-directed/random mutagenesis, DNA shuffling of homologous extant sequences, or the REAP approach.
Figure 2.
The use of phylogenetics for directed evolution and protein engineering. This schematic shows how variation contained within homologous sequences is captured by different directed evolution approaches and how this relates to sampling of sequence/function space. Branch lengths are not to scale. A, Site-directed mutagenesis approach randomly inserts mutations into the parent sequence. B, Standard DNA shuffling approach builds libraries that incorporate homologous sequence information from all branches of a phylogeny because the approach uses only extant (modern) sequence information. C, REAP approach builds libraries that incorporate homologous sequence information from only those branches of the phylogeny inferred to have undergone functional adaptation and divergence.
When site-directed mutagenesis is employed, a parent protein sequence is identified and mutated to generate the variant library. This technique may not rely on evolutionary knowledge and may result in dense sampling in the immediate sequence space of the parent protein even when structural and/or biochemical information is available (Figure 2A, right). While this approach is straightforward and works well when the desired function can be found in a sequence highly related to the parent protein, this approach generally samples a very limited area of sequence space and can thus entirely miss the functional sequence space of interest. This approach also has several limitations due to the fact that proteins are metastable and the average (random) amino acid replacement is destabilizing rather than stabilizing, resulting in a large proportion of variant proteins being non-functional altogether (Taverna and Goldstein 2002). We expect the REAP approach, and others that consider extant sequence information, to circumvent this problem because it only considers DNA substitutions and/or amino acid replacements that have been accepted by natural selection. This assumes that the sequence background is relatively robust to change and that epistatic effects are minimal (Harms and Thornton 2010). However, as certain phenotypic shifts have been shown to require a particular ancestral background, it may sometimes be necessary to introduce additional residues ancestral to both functional branches in order to create a permissive environment for functional shifts (Bridgham et al. 2009). This is based on the observation that particular combinations of ancestral states can have destabilizing effects through epistatic interactions.
Another approach commonly used is the DNA shuffling technique whereby amino acids present in modern sequences are combinatorially shuffled or recombined to generate a library. As seen in Figure 2B, patterns of amino acid residues that evolved either within a subfamily (branches bound by boxes) or along the branches that gave rise to the individual subfamilies (circled branches) are integrated during library design. Note that certain amino acid patterns observed in modern proteins arose within the subfamilies (boxed branches) and thus probably have little to offer in terms of generating novel biomolecular properties. These amino acid patterns arose mostly via neutral evolution assuming a lack of selective pressure to diversify within a given subfamily. Meaning, for example, that all proteins within the red family have equivalent emission spectra and the residues that differ between the five red-emitting proteins may not be useful when designing a library. The DNA shuffling approach will sample a large area of sequence space but may result in an intractably large library of variants to screen (Figure 2B, right).
Unlike the above standard approaches, the REAP method is based on explicit models of molecular evolution that attempt to eliminate amino acid patterns predicted to have minimal contributions to novel biomolecular functions. This is achieved by incorporating only the amino acid patterns that arose during the adaptive evolution of unique properties compared to the last common ancestor of the fluorescent proteins (Figure 2C, circled branches), and neglecting the amino acid patterns that arose within a family. In doing so, this increases or at least maintains the unique behaviors captured using the standard DNA shuffling approach, while limiting the number of mutations by culling those that are inferred to have minimal impact on functional diversification.
The REAP Method
General Methodology
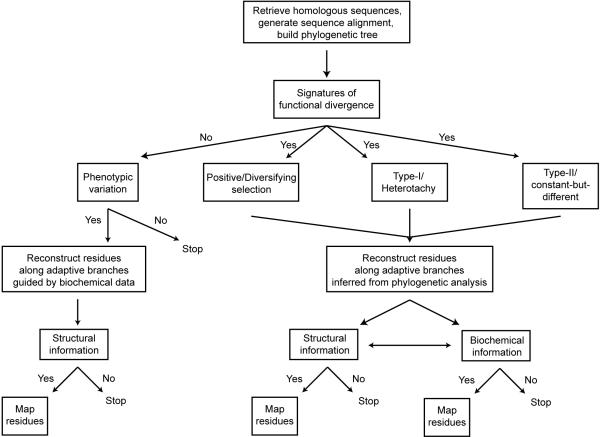
A general flow-chart for the REAP approach that can be used to guide variant library design is presented in Figure 3. The first step is to collect homologous sequences of a parent protein from databases such as NCBI or PFAM. A multiple sequence alignment is then created using software such as ClustalW (Larkin et al. 2007) or T-Coffee and manually inspected and refined as needed to obtain a trustworthy alignment. This alignment is used as input for a phylogenetic analysis to determine the relationships and evolutionary distances between the parent protein and its homologs. Software such as MrBayes (Huelsenbeck et al. 2001) can be used to construct a phylogenetic tree, which can be checked against existing knowledge of evolutionary relationships between the included species and adjusted if necessary.
Figure 3.
Flowchart for the REAP approach. The implementation of the REAP approach begins by collecting and aligning sequences from a protein family. A phylogenetic tree must then be constructed to capture the evolutionary relationship and distance between homologs. From this information, molecular models are used to detect functional divergence along branches of the phylogeny. The computational reconstruction of ancestral states of the protein along these branches is then used to identify residues and amino acids associated with the functional divergence. This will result in a list of candidate residues/mutations that may affect the function of the protein. This list of candidates can be further reduced if needed by incorporating known structural or biochemical information about the protein. For example, residues may be selected based on their proximity to the protein's active site. The final candidate residue list is then used to design the variant library to screen for the desired function. In this manner, the REAP approach results in a small number of residues/mutations to vary in the sequence library.
The phylogenetic tree and multiple sequence alignment are then used as input into software programs such as DIVERGE (Gu and Vander Velden 2002) and Rate Shift Analysis Server (Knudsen and Miyamoto 2001) that use evolutionary models to describe the replacements of amino acids, rate heterogeneity among sites, etc.. When these models detect functional divergence along branches of the phylogeny, ancestral sequence reconstruction, using programs such as PAML (Yang 2007), can be used to identify the specific residues that are changing along these branches. This list of residues can be further culled, as other directed evolution approaches often do, using protein structural models or biochemical information to direct mutations to particular sites or domains of a protein (Perez-Jimenez et al. 2009; Yuen and Liu 2007). When signatures of functional divergence are not detected, but there is known phenotypic variation among homologs, ancestral sequence reconstruction can still be used to provide a list of substitutions observed in the evolutionary history of the protein but these substitutions cannot be culled according to their connection to functional divergence.
REAP Analysis of family A DNA polymerases
DNA polymerases are routinely used for basic research and biotechnology applications. Although it is common practice to use standard nucleoside triphosphates as substrates for these polymerases, there is a growing interest in identifying/evolving/engineering DNA polymerases capable of incorporating non-standard nucleosides (Henry and Romesberg 2005; Patel et al. 2001; Sismour et al. 2004). For instance, incorporating non-standard nucleosides would allow researchers to explore alternative sugar-ring structures and modified backbone linkages in DNA, expand the information capacity of DNA beyond the four standard nucleosides, and develop novel sequencing technology. One of the earliest and most influential developments along the latter line included the engineering of Thermus aquaticus DNA polymerase (Taq) to accept dideoxynucleoside triphosphates. These modified nucleosides serve as non-reversible terminators for Sanger-based cycle sequencing and revolutionized DNA sequencing. Next-generation sequencing technology, such as sequencing-by-synthesis (SbS), will require polymerases that incorporate reversible terminators. Nucleosides whose 3' OH group, for instance, is replaced with an ONH2 group will block extension until the O-N bond in the ONH2 group is cleaved to restore the 3' OH and allow extension to proceed. The REAP approach was applied to Taq polymerase and its homologous family members (Family A DNA polymerases) to identify sites involved in expanded substrate recognition in order to engineer polymerases capable of incorporating dNTP- ONH2 reversible terminators.
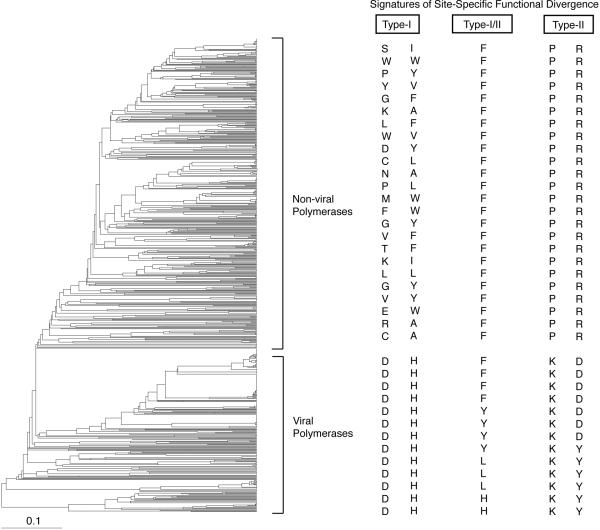
A phylogenetic tree and multiple sequence alignment of Family A DNA polymerases from eukarya, archaea, bacteria and viruses, were composed of 719 family A polymerase sequences available in the PFAM database at the time (PF00476) (Figure 4) (Bateman et al. 2004). Type-I and Type-II functional divergence was detected by feeding the alignment (with some taxa removed to reduce computational complexity) into DIVERGE and Rate Shift Analysis Server (http://www.daimi.au.dk/~compbio/rateshift/protein.html). The computational phylogenetic analysis of sequence information confirmed what was already known from the literature: functional divergence of polymerase behaviors has occurred along branches of the phylogeny separating viral and non-viral polymerases (Horlacher et al. 1995; Leal et al. 2006; Sismour et al. 2004; Tabor and Richardson 1995). Based on the observation that viral polymerases are better able to accept modified nucleosides than non-viral polymerases, we reasoned that extracting specific sequence information (sites responsible for functional divergence) from viral polymerases and placing them within the genetic background of the non-viral Taq polymerase would generate evolution-guided engineered polymerases with modified substrate specificities.
Figure 4.
Phylogeny of Family A DNA polymerases. The viral and non-viral clades used for REAP analysis are highlighted. Scale bar represents amino acid replacements/site/unit evolutionary time. Examples of patterns of types I & II functional divergence are also shown.
Amino acid residues replaced along the branches separating viral and non-viral polymerases were inferred using PAML (version 4.1) by incorporating the WAG matrix with rate variation following a gamma distribution. These analyses identified numerous sites as potentially involved in functional divergence between viral and non-viral polymerases both inside and outside the active-site cleft of the polymerase structure. We elected to focus our analysis on sites within the active-site only since these are known to alter substrate specificity (Henry and Romesberg 2005). A total of 57 amino acid replacements distributed across 35 sites were predicted to have the potential to expand the substrate recognition of Taq polymerase (Figure 5) and are listed in Table 1.
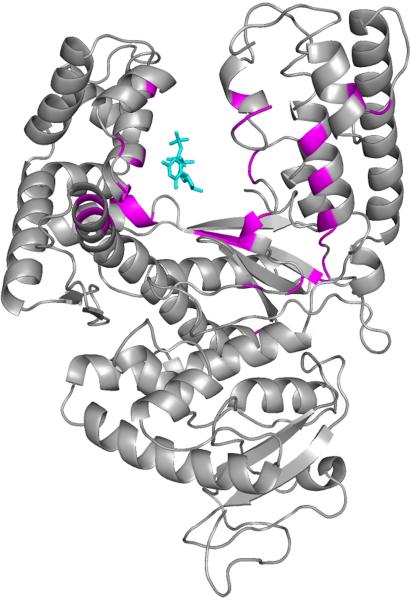
Figure 5.
Distribution of functional divergence sites mapped onto the structure of Taq polymerase. Locations of the 35 sites in Taq polymerase identified by the REAP analysis and mapped onto the polymerase structure and colored in magenta (PDB accession 5KTQ). The incoming nucleoside triphosphate substrate is shown in cyan.
Table 1.
Amino acid replacements identified by a REAP analysis of branches separating viral and non-viral lineages of Family A DNA polymerases
| Site | wild-type Taq | engineered Taq variant |
|---|---|---|
| 483 | Asn | Arg |
| 489 | Gln | His |
| 513 | Ser | Ile |
| 514 | Thr | Val |
| 520 | Glu | Ile, Gly |
| 536 | Arg | Ile |
| 540 | Lys | Ile |
| 544 | Thr | Ala |
| 545 | Tyr | Glu |
| 576 | Ser | Glu, His |
| 578 | Asp | Phe, Thr |
| 582 | Gln | Ala |
| 583 | Asn | Gln, Ser |
| 586 | Val | Lys |
| 587 | Arg | Val |
| 597 | Ala | Thr, Cys |
| 598 | Phe | Val, Trp |
| 600 | Ala | Ser |
| 604 | Trp | Gly |
| 608 | Ala | Gly, Lys, Glu |
| 609 | Leu | Cys, Pro, Ser |
| 610 | Asp | Trp |
| 614 | Ile | Glu, Gly, Gln |
| 615 | Glu | Ile |
| 616 | Leu | Ala, Ile, Asp |
| 625 | Asp | Ser, Leu, Ala |
| 660 | Arg | Asp |
| 667 | Phe | Tyr, His, Leu |
| 671 | Tyr | Phe |
| 673 | Met | Gly, Ala |
| 742 | Glu | Pro, Arg |
| 743 | Ala | Ser, Arg |
| 745 | Glu | His, Val |
| 746 | Arg | Ala |
| 777 | Ala | His |
NOTE - Sites are numbered according to wild-type Taq polymerse
Identification of polymerase variants from the REAP-designed library
From the REAP analysis a library of 93 Taq polymerase variants was designed where each variant had 3–4 amino acid replacements and each replacement was present in six of the variants (Chen et al. 2010). Each variant was cloned, expressed and then assayed for the ability to incorporate the reversible terminator dNTP-ONH2. Thirty variants (32%) were able to incorporate the reversible terminator to some degree and of these, eight of them were able to incorporate the reversible terminator with a threshold n+1 extension efficiency of at least 50% in two minutes. Two of the eight variants were exceptional in their ability to incorporate the modified nucleoside and were thus used in further assays to demonstrate their utility for sequencing reactions using reversible terminators (full details provided in Chen et al. 2010)
Our previous study of DNA polymerases clearly demonstrates the power of the REAP method. Using the REAP approach, a small number of amino acid replacements were identified as potentially useful to expand protein function, in this case the ability of polymerase to accept a non-standard nucleoside substrate. A small library of variants was then generated that could be assayed in a low-throughput manner to accurately screen for the desired function. A high percentage of the engineered proteins had a detectable increase in the desired function and two variants had a level of function high enough for use in the desired application without further modifications. Thus, the REAP approach exploited evolutionary information and models of molecular evolution to efficiently design and identify a protein with new functionality.
Discussion
Evolution is defined as change (mutation) in the heritable information (usually DNA) of biological systems from generation to generation. Mutations can be the result of either natural selection (i.e., adaptive) or genetic drift (neutral, or slightly deleterious), and occur randomly (Kimura 1991). In its purest form, experimental evolution of a protein in the laboratory would follow an analogous path; random changes followed by selection of variants for a particular property. The expanse of mutation space and limitations in selection schemes, however, render this approach impractical. To overcome these problems, researchers often focus their attention to particular positions and a subset of mutations to direct the evolution of a parent protein (Tobin et al. 2000; Van Regenmortel 2000).
The input for such direction varies considerably from in silico thermodynamic and steric structural considerations to in vitro mutagenesis experiments, and even activity profiles from initial rounds of directed evolution experiments (Fox et al. 2003; Fox et al. 2007; Korkegian et al. 2005; Saraf et al. 2004; Voigt et al. 2001). One of the most widely used approaches academically and commercially though involves shuffling of genetic differences between a parent protein and homologous members of the parent protein's evolutionary family (Crameri et al. 1998; Crameri et al. 1996). The success of 'DNA shuffling' (Molecular Breeding) is threefold: 1) sampling of sequence space is restricted, 2) library variants can contain sufficient diversity to generate novel biomolecular functions and 3) mutations contained within the set of homologs have already been subjected to evolutionary forces and are thus either adaptive, neutral or only slightly deleterious. This last point is noteworthy since it implies that none of the individual mutations extracted from the set of homologous sequences are deleterious enough to inactivate the protein. This assumption is, of course, incorrect if some of the homologs underwent pseudogenization or if particular mutations are context-dependent, beneficial or neutral in some sequence contexts but deleterious in others (Wang and Pollock 2005).
Although the shuffling approach greatly reduces the sequence space that is explored compared to randomly mutating a sequence, the approach scales exponentially with the number of homologs and the amount of sequence diversity contained within them. This restricts the amount of homologous sequence information that can be exploited to direct the evolution of a protein and thus decreases the chances of incorporating mutations that generate diversifying biomolecular functions. In addition, genome sequencing projects are rapidly identifying homologs for all gene families. This additional sequence information can be a burden for standard shuffling approaches, but it is favorable for approaches that exploit evolutionary information in designing libraries.
The ability to place large numbers of homologous sequences within an evolutionary framework provides an opportunity to determine whether conservation and variation are the result of functional divergence/constraint or common ancestry (Govindarajan et al. 2003; Lichtarge et al. 1996). Conserved sites are more likely to be associated with functional constraints if the evolutionary distance separating the sequences is long rather than short. Conversely, variable sites are more likely to be associated with functional divergence when they occur along short evolutionary paths (i.e., branches).
Several methods have been developed to identify functionally important sites within proteins based on evolutionary analysis of homologous sequences. For instance, ConSurf (Landau et al. 2005), uses multiple sequence alignments to score each residue for overall conservation amongst homologs. While this is a powerful way to identify sites implicated in the basic function of a protein, it is not able to identify sites associated with functional divergence in some protein subfamilies. One approach that can identify sites important for specific sub-family functionalization is Evolutionary Trace (Lichtarge, Bourne and Cohen 1996), which scores residues for conservation across all species and across consecutively smaller sub-classes. However, this method treats all mutations equally and does not use explicit evolutionary models to identify functionally relevant sites, thus missing certain types of sequence signatures of functional divergence. Two notable methods that attempt to harness the power of ancestral sequence reconstruction are Substitution Mapping (Skovgaard et al. 2006) and Ancestral Mutation (Yamashiro et al. 2010). Substitution mapping identifies sites of interest within a homolog that exhibit a desired functional quality by sequence comparison and then inserts these substitutions into a parent sequence. This approach relies on a priori knowledge of protein function and is thus not applicable to studies where the functions of various homologs have not been determined. The Ancestral Mutation method, on the other hand, does not rely on functional knowledge but rather on the fact that ancestral proteins often have increased thermostability. Thus, this method has been used to improve thermostability of enzymes by introducing ancestral residues into extant sequences.
REAP is unique from these other methods in that it uses explicit models of molecular evolution to identify sequence signatures of functional divergence within protein subfamilies. This evolution-guided strategy incorporates only those mutations inferred to be associated with new biomolecular properties during the evolution of a protein family, and to exclude the much larger set of mutations that do not lead to new functions (neutral or slightly deleterious mutations). For instance, assuming a background mutation rate of ca. 1×10−10 to 5×10−9 per base-pair per generation in E. coli (Lenski et al. 2003), neutral mutations will outnumber adaptive mutations 70–2000:1 per genome per generation (Perfeito et al. 2007). Eliminating the majority of these neutral mutations allows one to create libraries whose variants display a high level of functional diversity while restricting the overall sequence diversity, yielding a small library with a high proportion of active members which, in turn, permits the use of reliable low-throughput assays guaranteed to screen/select for variants exhibiting specific functions.
While the REAP methodology can be a powerful approach for protein engineering or directed evolution experiments it is not predicted to be ideal for all library designs. The approach requires numerous homologous sequences to generate an articulated phylogeny. Further, the phylogeny needs to represent a family of sequences with diverse behaviors guided by functional divergence, otherwise the extracted amino acid patterns may not generate novel function.
However, when there is sufficient homologous sequences and functional divergence, REAP may have substantial advantages over traditional library designs. The approach is intended to incorporate information from diverse family members to create a highly active and functional library. There is also no requirement for information regarding protein structure or the mutability of sites (mutagenesis experiments) to guide the library design. Equally important is that REAP libraries contain an order-of-magnitude fewer variants than most other types of libraries, allowing researchers to save time, money and to use low-throughput assays.
The general utility of the REAP approach will ultimately be determined by its ability to generate diverse biomolecules having novel functions. The approach has already been proven effective for the design of DNA polymerases capable of accepting non-standard nucleosides but further validation of the technique is needed for a wide array of protein designs. For now, we anticipate that the REAP approach will make substantial contributions to protein engineering and synthetic biology. For example, REAP could be used to generate protein variants capable of supporting unnatural amino acid incorporation during protein synthesis. The resulting biopolymers will then serve as the information (novel coding systems) and catalytic (novel side-chain chemistry) components of an expanded biology that we have termed “evolutionary synthetic biology” (Gaucher 2007).
Acknowledgements
This work was supported by a National Institutes of Health grant to EAG. MFC was supported by an NIH NRSA award and in part by the Emory University Fellowship in Research and Science Teaching (FIRST) program's NIH/NIGMS IRACDA grant number K12 GM000680-11. This work was also supported by the National Aeronautics and Space Administration's Exobiology and Astrobiology Programs.
Literature Cited
- Arnold FH, Georgiou G. Directed Enzyme Evolution: Screening and Selection Methods. Humana Press; Totowa, New Jersey: 2003. [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner SA, Gaucher EA. Evolution, language and analogy in functional genomics. Trends Genet. 2001;17:414–8. doi: 10.1016/s0168-9525(01)02320-4. [DOI] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol. 2004;59:121–32. doi: 10.1007/s00239-004-2597-8. [DOI] [PubMed] [Google Scholar]
- Brakmann S. Discovery of superior enzymes by directed molecular evolution. Chembiochem. 2001;2:865–71. doi: 10.1002/1439-7633(20011203)2:12<865::AID-CBIC865>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–9. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Gaucher EA, Leal NA, Hutter D, Havemann SA, Govindarajan S, Ortlund EA, Benner SA. Reconstructed evolutionary adaptive paths give polymerases accepting reversible terminators for sequencing and SNP detection. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2010;107:1948–1953. doi: 10.1073/pnas.0908463107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–91. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–9. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- Fox R, Roy A, Govindarajan S, Minshull J, Gustafsson C, Jones JT, Emig R. Optimizing the search algorithm for protein engineering by directed evolution. Protein Eng. 2003;16:589–97. doi: 10.1093/protein/gzg077. [DOI] [PubMed] [Google Scholar]
- Fox RJ, Davis SC, Mundorff EC, Newman LM, Gavrilovic V, Ma SK, Chung LM, Ching C, Tam S, Muley S, Grate J, Gruber J, Whitman JC, Sheldon RA, Huisman GW. Improving catalytic function by ProSAR-driven enzyme evolution. Nat Biotechnol. 2007;25:338–44. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- Gaucher EA. Ancestral sequence reconstruction as a tool to understand natural history and guide synthetic biology: Realizing (and extending) the vision of Zukerkandl and Pauling. Oxford University Press; 2007. pp. 20–33. [Google Scholar]
- Gaucher EA, Das UK, Miyamoto MM, Benner SA. The crystal structure of eEF1A refines the functional predictions of an evolutionary analysis of rate changes among elongation factors. Molecular Biology And Evolution. 2002;19:569–573. doi: 10.1093/oxfordjournals.molbev.a004113. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451:704–7. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Gu X, Miyamoto MM, Benner SA. Predicting functional divergence in protein evolution by site-specific rate shifts. Trends In Biochemical Sciences. 2002;27:315–321. doi: 10.1016/s0968-0004(02)02094-7. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Miyamoto MM, Benner SA. Function-structure analysis of proteins using covarion-based evolutionary approaches: Elongation factors. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2001;98:548–552. doi: 10.1073/pnas.98.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Miyamoto MM, Benner SA. Evolutionary, structural and biochemical evidence for a new interaction site of the leptin obesity protein. Genetics. 2003;163:1549–1553. doi: 10.1093/genetics/163.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan S, Ness JE, Kim S, Mundorff EC, Minshull J, Gustafsson C. Systematic variation of amino acid substitutions for stringent assessment of pairwise covariation. J Mol Biol. 2003;328:1061–9. doi: 10.1016/s0022-2836(03)00357-7. [DOI] [PubMed] [Google Scholar]
- Gu X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol Biol Evol. 2001;18:453–64. doi: 10.1093/oxfordjournals.molbev.a003824. [DOI] [PubMed] [Google Scholar]
- Gu X, Vander Velden K. DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics. 2002;18:500–1. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Thornton JW. Analyzing protein structure and function using ancestral gene reconstruction. Curr Opin Struct Biol. 2010;20:360–6. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry AA, Romesberg FE. The evolution of DNA polymerases with novel activities. Curr Opin Biotechnol. 2005;16:370–7. doi: 10.1016/j.copbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Horlacher J, Hottiger M, Podust VN, Hubscher U, Benner SA. Recognition by viral and cellular DNA polymerases of nucleosides bearing bases with nonstandard hydrogen bonding patterns. Proc Natl Acad Sci U S A. 1995;92:6329–33. doi: 10.1073/pnas.92.14.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Evolution - Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution: a review of recent evidence. Jpn J Genet. 1991;66:367–86. doi: 10.1266/jjg.66.367. [DOI] [PubMed] [Google Scholar]
- Knudsen B, Miyamoto MM. A likelihood ratio test for evolutionary rate shifts and functional divergence among proteins. Proc Natl Acad Sci U S A. 2001;98:14512–7. doi: 10.1073/pnas.251526398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkegian A, Black ME, Baker D, Stoddard BL. Computational thermostabilization of an enzyme. Science. 2005;308:857–60. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Research. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leal NA, Sukeda M, Benner SA. Dynamic assembly of primers on nucleic acid templates. Nucleic Acids Res. 2006;34:4702–10. doi: 10.1093/nar/gkl625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman N, Unrau PJ. Recombination during in vitro evolution. Journal of Molecular Evolution. 2005;61:245–252. doi: 10.1007/s00239-004-0373-4. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Winkworth CL, Riley MA. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J Mol Evol. 2003;56:498–508. doi: 10.1007/s00239-002-2423-0. [DOI] [PubMed] [Google Scholar]
- Liao J, Warmuth MK, Govindarajan S, Ness JE, Wang RP, Gustafsson C, Minshull J. Engineering proteinase K using machine learning and synthetic genes. BMC Biotechnol. 2007;7:16. doi: 10.1186/1472-6750-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–58. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H. Heterotachy, an important process of protein evolution. Mol Biol Evol. 2002;19:1–7. doi: 10.1093/oxfordjournals.molbev.a003973. [DOI] [PubMed] [Google Scholar]
- Lutz S, Patrick WM. Novel methods for directed evolution of enzymes: quality, not quantity. Curr Opin Biotechnol. 2004;15:291–7. doi: 10.1016/j.copbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Ness JE, Cox AJ, Govindarajan S, Gustafsson C, Gross RA, Minshull J. Empirical biocatalyst engineering: escaping the tyranny of high throughput screening. American Chemical Society; Washington, DC: 2005. [Google Scholar]
- Ness JE, Kim S, Gottman A, Pak R, Krebber A, Borchert TV, Govindarajan S, Mundorff EC, Minshull J. Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently. Nat Biotechnol. 2002;20:1251–5. doi: 10.1038/nbt754. [DOI] [PubMed] [Google Scholar]
- Patel PH, Kawate H, Adman E, Ashbach M, Loeb LA. A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity. J Biol Chem. 2001;276:5044–51. doi: 10.1074/jbc.M008701200. [DOI] [PubMed] [Google Scholar]
- Perez-Jimenez R, Li JY, Kosuri P, Sanchez-Romero I, Wiita AP, Rodriguez-Larrea D, Chueca A, Holmgren A, Miranda-Vizuete A, Becker K, Cho SH, Beckwith J, Gelhaye E, Jacquot JP, Gaucher EA, Sanchez-Ruiz JM, Berne BJ, Fernandez JM. Diversity of chemical mechanisms in thioredoxin catalysis revealed by single-molecule force spectroscopy (vol 16, pg 890, 2009) Nature Structural & Molecular Biology. 2009;16:1331–1331. doi: 10.1038/nsmb.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L, Fernandes L, Mota C, Gordo I. Adaptive mutations in bacteria: high rate and small effects. Science. 2007;317:813–5. doi: 10.1126/science.1142284. [DOI] [PubMed] [Google Scholar]
- Pupko T, Galtier N. A covarion-based method for detecting molecular adaptation: application to the evolution of primate mitochondrial genomes. Proc Biol Sci. 2002;269:1313–6. doi: 10.1098/rspb.2002.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraf MC, Horswill AR, Benkovic SJ, Maranas CD. FamClash: a method for ranking the activity of engineered enzymes. Proc Natl Acad Sci U S A. 2004;101:4142–7. doi: 10.1073/pnas.0400065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sismour AM, Lutz S, Park JH, Lutz MJ, Boyer PL, Hughes SH, Benner SA. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1. Nucleic Acids Research. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard M, Kodra JT, Gram DX, Knudsen SM, Madsen D, Liberles DA. Using evolutionary information and ancestral sequences to understand the sequence-function relationship in GLP-1 agonists. Journal of Molecular Biology. 2006;363:977–988. doi: 10.1016/j.jmb.2006.08.066. [DOI] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc Natl Acad Sci U S A. 1995;92:6339–43. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna DM, Goldstein RA. Why are proteins marginally stable? Proteins-Structure Function and Genetics. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nature Reviews Genetics. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- Tobin MB, Gustafsson C, Huisman GW. Directed evolution: the 'rational' basis for 'irrational' design. Curr Opin Struct Biol. 2000;10:421–7. doi: 10.1016/s0959-440x(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel MH. Are there two distinct research strategies for developing biologically active molecules: rational design and empirical selection? J Mol Recognit. 2000;13:1–4. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<1::AID-JMR490>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Voigt CA, Mayo SL, Arnold FH, Wang ZG. Computational method to reduce the search space for directed protein evolution. Proc Natl Acad Sci U S A. 2001;98:3778–83. doi: 10.1073/pnas.051614498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Spencer M, Susko E, Roger AJ. Testing for covarion-like evolution in protein sequences. Mol Biol Evol. 2007;24:294–305. doi: 10.1093/molbev/msl155. [DOI] [PubMed] [Google Scholar]
- Wang ZO, Pollock DD. Context dependence and coevolution among amino acid residues in proteins. Methods Enzymol. 2005;395:779–90. doi: 10.1016/S0076-6879(05)95040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–51. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro K, Yokobori S, Koikeda S, Yamagishi A. Improvement of Bacillus circulans beta-amylase activity attained using the ancestral mutation method. Protein Eng Des Sel. 2010;23:519–28. doi: 10.1093/protein/gzq021. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology And Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Kumar S, Nei M. A New Method of Inference of Ancestral Nucleotide and Amino-Acid-Sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Arnold FH. Directed evolution of subtilisin E in Bacillus subtilis to enhance total activity in aqueous dimethylformamide. Protein Eng. 1996;9:77–83. doi: 10.1093/protein/9.1.77. [DOI] [PubMed] [Google Scholar]
- Yuen CM, Liu DR. Dissecting protein structure and function using directed evolution. Nature Methods. 2007;4:995–997. doi: 10.1038/nmeth1207-995. [DOI] [PMC free article] [PubMed] [Google Scholar]