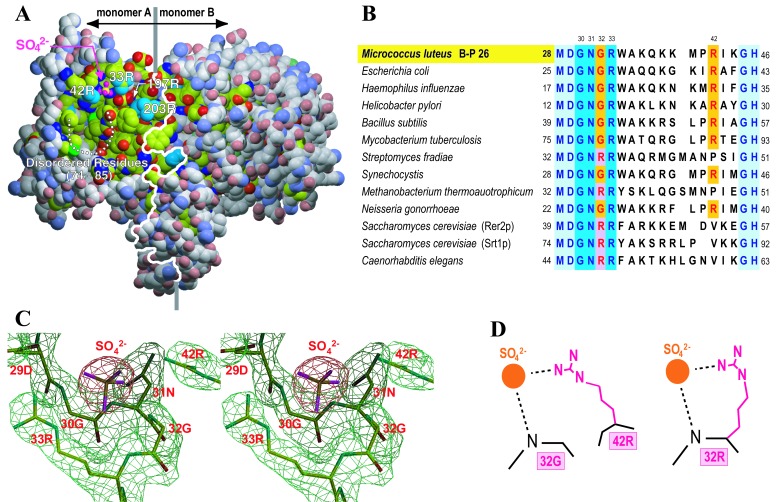
Figure 3.
Active site in the UPS structure. (A) Space filling model of the UPS dimer. C atoms are indicated in yellowish green and gray, in which the former color corresponds to conserved residues and the latter nonconserved residues among the cis-prenyltransferase family. These conserved residues form a large hydrophobic cleft. N, O, and S atoms are colored in blue, red, and green, respectively. The white line indicates the interface of the dimer, where the left and right monomers are named A and B. White dotted line shows the disordered residues from 74S to 85V. This figure was prepared in the same manner as Fig. 2 A and B. (B) A comparison of amino acid sequences in 13 proteins that have sequence homology with UPS from M. luteus B-P 26 (12, 13). Only residues corresponding to 28–46 of M. luteus UPS are presented. The proteins from M. luteus, E. coli, and Hemophilus influenzae demonstrate UPS activity (10, 12, 13). RER2p from S. cerevisiae has cis-prenyltransferase activity (11). The others are potential cis-prenyltransferases homologous with M. luteus UPS. (C) Stereo view of the electron density map around the structural P-loop motif and the sulfate ion. The final refined structure is superimposed. (D) Three-dimensionally conserved guanidinium head of arginine. The positively charged guanidinium group of either 42R (in the case of 32G; Left) or 32R (in the case of 32R; Right) binds to the diphosphate head of the substrate.