Figure 1.
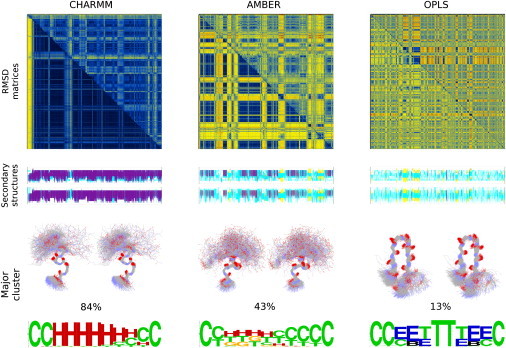
Stability, secondary structures, and major clusters. Direct comparison among the RMSD matrices (top row), the secondary structure assignments (middle row), and the prominent structures obtained from each of the three force fields. For both the RMSD matrices and the secondary structure assignments, the horizontal axes correspond to simulation time and range from zero to 1.3 μs for CHARMM, 2.9 μs for AMBER, and 3.4 μs for OPLS. The RMSD matrices (top row) have been calculated using either all Cα atoms (upper half of the matrices), or only those of residues 1–7 (lower half). For all three matrices, the origin is at the top left-hand corner and the linear color scale ranges from dark blue (corresponding to an RMSD of zero), through yellow (for intermediate RMSDs), to dark red (large RMSDs). For the lower halves of the matrices the RMSDs range from 0.0 to 6.2 Å, becoming 0.0 to 10.9 Å when using all Cα atoms (upper halves). For the secondary structure assignments, both the DSSP (upper graph) and STRIDE-derived assignments (lower graph) are shown. (For both graphs, the color code is: α-helix → magenta, 310-helix → red, β-structure → yellow, turns → cyan, and coil → white.) The third row shows the structures (wall-eyed stereodiagrams), the frequencies, and the secondary structure assignments (in the form of a weblogo) for the major clusters obtained from each trajectory. For the structure diagrams only backbone atoms are shown to reduce clutter. For the weblogo, H is α-helix, G is 310-helix, C is coil, T is turn, E is extended structure, and B is bridge as reported by the program STRIDE. Calculation of RMSD matrices and cluster analysis was performed with the program CARMA.
