Figure 2.
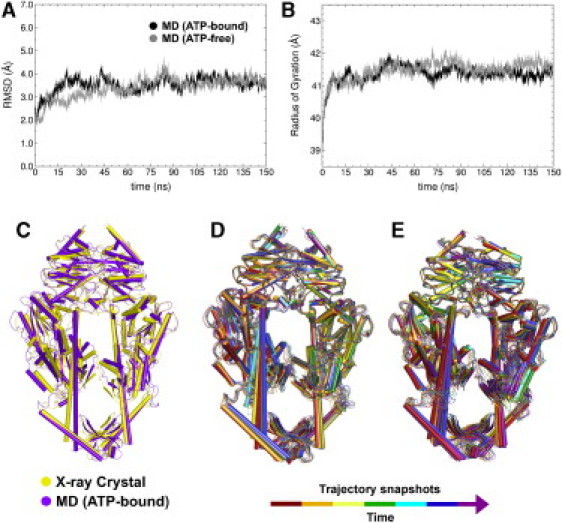
(A) RMSD of protein and DNA backbone atoms with respect to the first simulation snapshot for MutS⋅DNA(+T)⋅ATP (black) and MutS⋅DNA(+T) (gray) simulations. (B) Radius of gyration of protein backbone for the corresponding simulations. (C) Crystal structure of MutS⋅ADP⋅BeF3− (PDB code 1nne) (yellow) superimposed with the calculated average structure for MD on MutS⋅DNA(+T)⋅ATP (ATP-bound) (purple). (D and E) Simulation snapshots of ATP-bound (D) and ATP-free (E) complexes.
