Abstract
Sexual dimorphism (SD) in morphological, behavioural and physiological features is common, but the genetics of SD in the wild has seldom been studied in detail. We investigated the genetic basis of SD in morphological traits of threespine stickleback (Gasterosteus aculeatus) by conducting a large breeding experiment with fish from an ancestral marine population that acts as a source of morphological variation. We also examined the patterns of SD in a set of 38 wild populations from different habitats to investigate the relationship between the genetic architecture of SD of the marine ancestral population in relation to variation within and among natural populations. The results show that genetic architecture in terms of heritabilities, additive genetic variances and covariances (as well as correlations) is very similar in the two sexes in spite of the fact that many of the traits express significant SD. Furthermore, population differences in threespine stickleback body shape and armour SD appear to have evolved despite constraints imposed by genetic architecture. This implies that constraints for the evolution of SD imposed by strong genetic correlations are not as severe and absolute as commonly thought.
Keywords: body shape, G, Gasterosteus, genetic correlation, heritability, sexual dimorphism
Introduction
Sexual dimorphism (SD) in morphological, behavioural and physiological features is of common occurrence in the wild. SD in traits closely related to reproduction is typically interpreted in terms of adaptation of males and females to their respective reproductive roles (reviewed by, for example, Greenwood and Adams, 1987; Anderson, 1994; Short and Balabban, 1994; Mealey, 2000; Fairbairn et al., 2007), while SD in traits not directly related to reproduction has been suggested to be a result of adaptation of the two sexes to different ecological niches (Slatkin, 1984; Hedrick and Temeles, 1989).
The adaptive explanations for the evolution of SD in different contexts have been widely accepted, yet a largely unresolved issue lies beneath—the role of genomic conflict between the sexes, which is caused by the same genes determining the same traits in both sexes (Rice, 1984; Gibson et al., 2002; Bonduriansky and Rowe, 2005; Fairbairn et al., 2007). In other words, the genetic correlation between the sexes (rg(MF)) is likely to slow down the evolution of SD (Lande, 1980; Reeve and Fairbairn, 2001; Fairbairn et al., 2007), and theoretically, in the extreme case, when genetic correlation between the sexes for a given trait is perfect (rg(MF)=1), SD should not evolve. However, even in the presence of high rg(MF), SD can evolve if the sexes differ in the extent of genetic variance for a trait (Lynch and Walsh, 1998).
Simulations have shown that genetic correlations between the sexes constrain the evolution of SD much less than predicted by Lande's (1980) model (Reeve and Fairbairn, 2001), which is built around the matrix of genetic variances and covariances between the sexes. Evidence from empirical work is mixed, indicating that the issue is not fully resolved. Studies that have tested the association between rg(MF) and the degree of SD have both found (Ashman, 2003; Bonduriansky and Rowe, 2005; McDaniel, 2005; Fairbairn, 2007) and failed to find (Cowley et al., 1986; Cowley and Atchley, 1988; correlation reported in Fairbairn and Roff, 2006) significant negative correlation between these two measures. A survey five dioecious and gynodioecious plant species also came up with mixed results; the relationship between genetic correlation between the sexes and level of SD depended on the species in question (Ashman and Majetic, 2006). A recent meta-analysis of 66 dioecious plant and animal populations uncovered a negative correlation between rg(MF) and SD, indicating that rg(MF) more often than not constrains the evolution of SD (Poissant et al., 2010). The most likely reason for this is that rg(MF) of morphological traits are usually large and positive, while rg(MF) of physiological and fitness traits are typically smaller (Poissant et al., 2010).
SD in threespine stickleback (Gasterosteus aculeatus) body shape (Leinonen et al., 2006; Kitano et al., 2007; Aguirre et al., 2008; Spoljaric and Reimchen, 2008) and armour (Moodie, 1972; Moodie and Reimchen, 1976; Reimchen, 1980; Reimchen et al., 1985; Fernández et al., 2000; Reimchen and Nosil, 2006; Aguirre et al., 2008) has been reported from a number of wild populations across the species distribution range. The level of SD in threespine stickleback body shape has been shown to vary across habitats (Spoljaric and Reimchen, 2008), as well as over time in a single population (Aguirre et al., 2008). Although the genetic basis of the individual characters, especially body armour (for example, Peichel et al., 2001; Cresko et al., 2004; Schluter et al., 2004; Colosimo et al., 2005), is relatively well established, the genetic basis of SD has received less attention (but see Kitano et al., 2007; Albert et al., 2008). It has been shown that the majority of QTLs for body shape are located in the sex-determining region (Albert et al., 2008). It is also evident that ancestral SD reflects the body shape divergence observed between the limnetic and benthic ecotypes (Albert et al., 2008). These findings together suggest that ancestral SD has had a large role in promoting population divergence in the threespine stickleback species complex by providing variation on which selection can act (Aguirre et al., 2008; Albert et al., 2008).
The main aim of this study was to investigate the genetic basis of SD in a marine (ancestral) threespine stickleback population in detail. Specifically, we aimed to answer the following questions: (1) is the genetic basis of body shape and armour variation in males and females similar? If so, there should be no differences in sex-specific heritability estimates, or in the structure and orientation of sex-specific genetic covariance matrices among sexes reared under standard environmental conditions. (2) Do the possible similarities in genetic architecture of the traits in question impose constraints on the evolution of SD? A negative association between rg(MF) and the level of SD across different traits would indicate that the genomic conflict constrains the evolution of SD. If the constraints are severe and the genomic conflict has not been resolved, we should not find differences in the levels of SD in wild populations from different habitats, which leads to the final question. (3) Is the genetic basis of SD, as inferred from the laboratory data, reflected in the patterns of variation in SD among wild populations?
Materials and methods
Sampling and fish rearing
A total of 1919 fish from 37 wild populations in Europe and 1 in the United States (Maine, Penebscot Bay) were sampled (Supplementary Table S1). The populations were classified as sea (n=13), lake (n=4), pond (n=10), migratory (n=3) and resident river (n=8) according to their sampling sites (for phylogenetic relationships of these populations see: Mäkinen et al., 2006; Mäkinen and Merilä, 2008).
For the lab cross, we used a population from the Baltic Sea (Vuosaari, Helsinki; 60°10′N, 25°00′E) as a representative ancestral population. Previous studies have shown that this population differs very little from oceanic populations, both genetically (Mäkinen et al., 2006) and phenotypically (Leinonen et al., 2006) and is likely to represent the ancestral form of the Fennoscandian and Northern European populations (Münzing, 1963; Reusch et al., 2001; Leinonen et al., 2006; Mäkinen et al., 2006).
Mature male and gravid female threespine sticklebacks were collected during the breeding season in June 2006. A seine net with 6 mm mesh size was used for capture. The fish were transported into the aquacultural facility at the University of Helsinki, and the crosses were made in vitro immediately on arrival. The crosses were performed using a nested paternal half-sib design, that is, each male was crossed with two different females. In total, 42 males were crossed with 84 females. Males were anaesthetized and killed with an overdose of MS-222 (tricane methanesulphonate) before extracting the testes and finely chopping them in a few drops of water. The sperm solution was used to fertilize eggs, which were obtained by gently pressing the abdomen of the females. The fertilized eggs were placed in cylindrical plastic containers with a plastic mesh bottom. The containers were submerged in 10 l plastic tanks with air supply to keep the water saturated with oxygen. Throughout the experiment the water temperature was set to 17 °C and the photoperiod to 12-h light:12-h dark. Once the eggs hatched, each clutch was divided into two replicate 10 l plastic tanks, and fed daily in excess with Artemia nauplii. After 3 months, the fish were also fed daily with chopped chironomid larvae.
The number of sticklebacks in each replicate tank was set to 15. Families with <15 fish per replicate (11 out of 84 half-sib families) were pooled with other families to make the density in each tank 15 individuals. The fish from different families were marked with fluorescent elastomer (Northwest Marine Technology, Inc., Shaw Island, WA, USA) fibres before pooling. The fish were killed after 6 months, when they had reached ca 4 cm in standard length and thus completed their lateral plate development (Hagen, 1973; Bell, 1981). The fish were fixed in 4% formalin and stored horizontally for a minimum of 1 month, and then stained with Alizarin Red S using a procedure that has been previously used by, for example, Pritchard and Schluter (2001). Before fixation, the pectoral fins were clipped and stored in 96% alcohol for sexing.
DNA from the fin samples were extracted using a method described by Duan and Fuerst (2001) and the sex was determined by amplifying a part of 3′UTR of IDH gene (Peichel et al., 2004). PCR-conditions were optimized in order to get the male band more visible. The final PCR reaction volume was 10 μl and consisted of 1 μl template DNA diluted 1:10, 1 × NH4 reaction buffer (Bioline), 1.5 m MgCl2 (Bioline, London, UK), 20 μ of each dNTP (Finnzymes, Espoo, Finland), 0.32 μ of IDH exon II 37F and IDH exon II 290R primers and 0.25 U of BioTaq polymerase (Bioline). PCR was conducted on thermal cycler MBS (Thermo, Waltham, MA, USA) according to following thermal profile: 94 °C for 3 min, 38 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min and the final extension at 72 °C for 5 min. Following the PCR, 5 μl of amplicon was run on a 2% Agarose LE (Cambrex, East Rutherford, NJ, USA) with 1 × loading dye (Fermentas, Helsinki, Finland). Fragment sizes were determined against size standard (GeneRuler, Fermentas). Males had two fragments, ∼280 bp and 300 bp, females only one, 300 bp. Each PCR-plate contained verified male-DNA as a positive control, as well as a negative control (no sample). Fish from the wild populations were sexed via gonad inspection.
Data acquisition
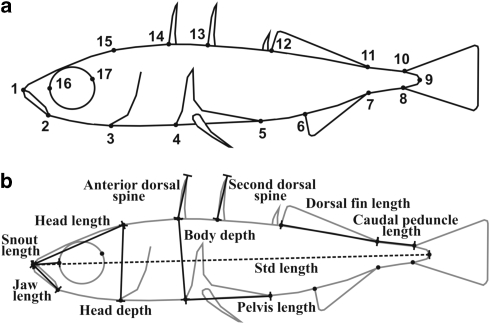
To measure body shape variation, the left side of each fish was photographed with a digital camera. All photographs were taken from a standard angle, and a ruler was placed in each photograph for scaling. Landmark-based geometric morphometrics were used to measure the shape of the specimens. On each fish, 17 homologous landmarks were digitized using tpsDig 2 Version 2.10 (Rohlf, 2006). The landmarks (Figure 1a; Supplementary Table S2) were selected such that they would capture overall body shape with as few variables as possible, while still describing the morphological features that have been shown to be variable in threespine stickleback populations from different habitats (for example, Walker, 1997; Leinonen et al., 2006; Aguirre et al., 2008; Sharpe et al., 2008). The lengths of the anterior and second dorsal spines were also measured from the photographs using tpsDig 2 Version 2.10 (Rohlf, 2006).
Figure 1.
Illustration of used landmarks. (a) Landmarks used for the body shape analyses (see Supplementary Table S2 for the description of the landmarks). (b) Linear measurements chosen based on the results of the body shape analysis.
Digitized landmarks were superimposed using tpsRelw Version 1.45 (Rohlf, 2007), which aligns, scales and rotates the landmark configurations using generalized orthogonal least squares superimposition with scaling to unit centroid size (Rohlf and Slice, 1990). The same program was used to calculate partial warp scores (Bookstein, 1991), which were used as shape variables in the subsequent analyses, and relative warp scores (which are essentially principal components, because equal weight was set for all partial warps at all spatial scales) for each individual. Linear measurements were calculated from distances between landmarks.
Although all the specimens were fixed, stained and photographed in a standardized manner, the possibility of measurement error because of distortion associated with the posture of the specimens and digitizing still remains (Arnqvist and Mårtensson, 1998). To minimize the possible effect of systematic error, the fish were photographed in random order. There is also the possibility of an arching effect because of the non-rigid structure of the fish (for example, Albert et al., 2008; Valentin et al., 2008). In previous studies of threespine stickleback body shape, the effect of bending on landmark data has been eliminated by removing the eigenvectors (with their associated eigenvalues) that have described vertical arching of the body (Albert et al., 2008; Sharpe et al., 2008). In our data, the first phenotypic eigenvector also included vertical arching, but since the first eigenvector possibly also includes some genetic effects, we decided to keep all the shape data for the subsequent quantitative genetic analyses. We tested the effect of removing the bending with the ‘unbend'—option implemented in tpsUtil (Ver. 1.38; Rohlf, 2006). The results of the analyses of genetic variance components with unbent specimens corresponded to those with the original specimens, and for the phenotypic data, the results of the unbent specimens corresponded to the results of the original specimens with the first principal component removed (results not shown).
Quantitative genetic analyses
Genetic and phenotypic variance components were estimated using restricted maximum likelihood as implemented in ASReml software (Gilmour et al., 2001). The variance components were estimated for partial warp scores (that is, shape variables) and body armour traits, as well as for linear measurements (calculated from interlandmark distances) that correspond to those body shape features that varied significantly in the geometric morphometric analyses (Figure 1b). Linear measurements were used to overcome statistical and computational issues caused by the high dimensionality of geometric morphometric data (see below). Using linear measurements also simplifies the biological interpretation of the covariances and correlations between body shape and armour traits. All the variance and covariance components were estimated using animal model, in which animal and dam were fitted as random effects. As each half-sib family was reared in a replicate block, variance due to block was fitted in all the models as a fixed effect. All the analyses were performed separately for males and females. Sex-specific narrow sense heritabilities (h2) were estimated for the linear measurements as well as for the armour traits. Heritabilites of SD (h2(SD)) of the linear measurements and lateral plate numbers were estimated using the formula from Chapuis et al. (1996):
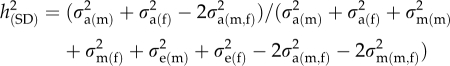 |
where SD is the difference in the sex specific means of a given trait, σ2a is the additive genetic, σ2m maternal and σ2e environmental variance component for the trait in males (m), females (f) or the covariance of the trait between the sexes (m,f). For the estimation of between-sex genetic correlations (rg(MF)), male and female measurements were considered as different traits. Spearman's rank correlation was used to test for the association between rg(MF) and the level of SD, which was quantified with the index: SD=(size of the larger sex/size of the smaller sex)–1 (Lovich and Gibbons, 1992).
The distribution of lateral plate numbers differed significantly from the probability distributions implemented in the ASReml software (for details, see Gilmour et al., 2006). The distribution of the lateral plates was closest to the negative binomial distribution, and we therefore used the variance components from the analysis fitted with a negative binomial distribution with a logit link function. Fitting different probability distributions did not significantly affect the heritability estimates for the lateral plates (results not shown). As a result of software limitations (that is, lack of convergence in models with more than two dependent variables) that were due to the high number of dimensions (30 for the shape variables, 10 for the linear measurements), it was not possible to estimate all the covariance components by including all the variables in the same model simultaneously. The G and P of body shape were therefore compiled from pairwise covariances between the partial warps. For the linear measurements, the genetic and phenotypic variance components, G matrices, genetic correlations and the directions of maximum variance were estimated similarly as for the landmark data; the only difference being that standard length (see Figure 1b) was used instead of centroid size as a covariate. The linear measurements were ln-transformed for the calculation of G matrices to remove the effects of absolute size of the traits on the variances. Genetic covariances and correlations between lateral plates and linear measurements were also estimated with standard length as a covariate, but because the distribution of the lateral plate numbers differed significantly from normality (that is, the distribution of the other traits), these covariances were not included in the comparisons of divergence vectors. The significance of the genetic (co)variances was tested with likelihood ratio tests, in which the full models were compared with those with (co)variances set to zero. The same method was used to test whether rg(MF) estimates differed from unity. Differences in heritabilities and variance components between the sexes were tested with paired t-tests.
Comparisons of G and P
To estimate the directions of greatest genetic covariance (gmax) for each sex, we performed principal component analyses of the G matrices, where the dominant eigenvectors represented gmax (Schluter, 1996). Accordingly, the dominant eigenvectors of the P matrices represented the maximum directions of phenotypic covariance (pmax). For the shape variables, the directions of genetic and phenotypic variances within and between the sexes were compared by vector correlations (which in essence are the inner products of the corresponding eigenvectors) and by calculating the angle between the vectors (arccosines of the vector correlations; Pimentel, 1979). Confidence intervals for the correlations between the dominant genetic and phenotypic eigenvectors were estimated by a randomization test, in which the observed vector correlations were compared against a null distribution that was created by performing vector correlations of 10 000 pairs of random vectors in 30-dimensional space (for example, Klingenberg and McIntyre, 1998; Klingenberg and Leamy, 2001).
Correlations between the directions of genetic and phenotypic covariances among and between the sexes for the linear measurements were calculated in the same way as for the partial warps, with the addition of calculating confidence intervals for the correlations by bootstrapping over families (cf. Schluter, 1996; McGuigan et al., 2005). This was done by creating 1000 replicates of the G and P by randomly sampling families with replacement, and from the dominant eigenvectors of principal component analyses on these replicates of G and P, random pairings were made to generate 1000 estimates of the correlations and their respective angles.
Results
Lab-reared fish
Trait means
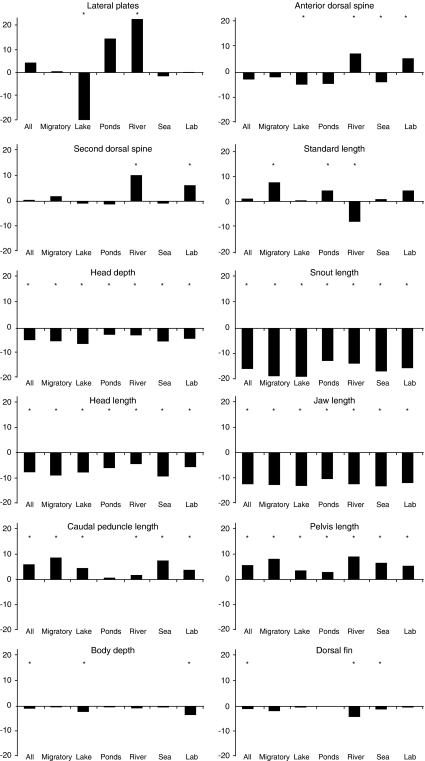
In total, 766 males and 1140 females from 84 half-sib families were successfully sexed. The main differences in body shape between the sexes in the laboratory cross were that males had relatively deeper bodies, larger heads, longer jaws and snouts, more posterior pelvises, and shorter but deeper caudal peduncles (Figure 2a). A multivariate analysis of variance on all the body shape variables confirmed significant SD in the laboratory cross (Wilks' λ=0.34, P<0.001). All the linear measurements differed significantly between the sexes, with the exception of the dorsal fin length and the number of lateral plates (Figure 3). In addition to the differences in body shape, the sexes differed also in relative dorsal spine length (females > males; Figure 3).
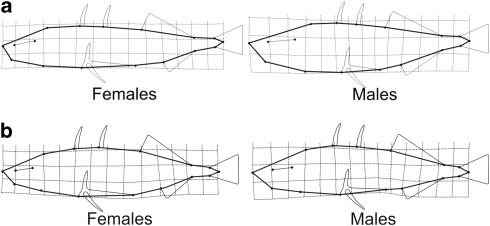
Figure 2.
Body shape differences between male and female threespine sticklebacks (a) in laboratory and (b) in the wild sea populations. The illustrations depict the mean shapes of females and males with differences five times exaggerated. Bending of the grid shows the extent of dimorphism in different parts of the body (that is, more bending, more differentiation). A square grid would represent the mean for all specimens.
Figure 3.
SD in different threespine stickleback populations expressed as a percentage of male mean. Traits that are larger in females have positive values, while trait means that are higher in males have negative values. Values significantly different from 0 at P<0.05 level are marked with an asterisk.
Heritabilities and genetic correlations
There were no significant differences in the heritability estimates between the sexes (Table 1). However, maternal effect variance components were significantly higher in females than males for the length of the first dorsal spine, snout length, head depth and body depth (Table 1). In contrast, males had a higher maternal effect variance component in the length of the caudal peduncle (Table 1). Heritabilities of SD were expectedly low, with body depth dimorphism having the highest estimated h2SD (Table 1). Genetic correlations between sexes were high, as could be expected, and none of them differed significantly from unity. With all the linear measurements combined, there was no significant correlation between rMF and the level of SD (rs=0.23, P=0.46).
Table 1. Genetic parameters (±s.e.) for the length measurements and lateral plates for the two sexes: heritability (h2), additive genetic variance (VA), maternal variance (VM) and residual variance (VR).
| Trait | rA | h2SD | Sex | h2 | VA (all values × 10−3) | VM (all values × 10−9, except*= × 10−3) | VR (all values × 10−3) |
|---|---|---|---|---|---|---|---|
| Caudal peduncle | 0.96±NA | 0.04 | Females | 0.46±0.08 | 1.28±0.26 | 0.28±0.031 | 1.48±0.17 |
| Males | 0.49±0.09 | 1.21±0.27 | 3.47±0.49 | 1.27±0.18 | |||
| Dorsal fin | 0.92±0.08 | 0.17 | Females | 0.69±0.09 | 3.05±0.56 | 0.09±0.020 | 1.40±0.31 |
| Males | 0.69±0.09 | 2.33±0.46 | 0.07±0.019 | 1.04±0.26 | |||
| Dorsal spine1 | 0.98±0.03 | 0.04 | Females | 0.66±0.09 | 1.35±0.25 | 1.10±0.23 | 0.69±0.14 |
| Males | 0.72±0.10 | 1.24±0.24 | 0.02±0.007 | 0.48±0.14 | |||
| Dorsal spine2 | 0.99±0.08 | 0.02 | Females | 0.63±0.09 | 1.16±0.22 | 0.02±0.004 | 0.69±0.13 |
| Males | 0.69±0.10 | 1.08±0.22 | 0.02±0.006 | 0.49±0.12 | |||
| Head depth | 1.00±NAa | 0.09 | Females | 0.47±0.08 | 0.37±0.08 | 0.02±0.003 | 0.41±0.05 |
| Males | 0.70±0.10 | 0.66±0.13 | 0.01±0.003 | 0.28±0.08 | |||
| Head length | 0.80±0.08 | 0.16 | Females | 0.41±0.07 | 0.84±0.17 | 0.06±0.006 | 1.19±0.12 |
| Males | 0.51±0.09 | 1.10±0.24 | 0.04±0.007 | 1.07±0.16 | |||
| Body depth | 0.90±0.09 | 0.28 | Females | 0.78±0.09 | 0.87±0.16 | 0.79±0.28 | 0.24±0.09 |
| Males | 0.76±0.10 | 1.03±0.20 | 0.02±0.007 | 0.32±0.11 | |||
| Jaw length | 0.95±0.14 | 0.08 | Females | 0.44±0.07 | 0.20±0.04 | 0.01±0.001 | 0.26±0.03 |
| Males | 0.56±0.09 | 0.29±0.06 | 0.01±0.002 | 0.23±0.04 | |||
| Pelvis length | 0.91±0.23 | 0.07 | Females | 0.48±0.08 | 1.33±0.28 | 3.18±0.38 | 1.43±0.17 |
| Males | 0.22±0.20 | 0.59±0.54 | 2.7*±2.5* | 1.84±0.30 | |||
| Snout length | 0.93±<0.0001 | 0.05 | Females | 0.46±0.08 | 0.33±0.07 | 0.02±0.003 | 0.38±0.04 |
| Males | 0.57±0.09 | 0.49±0.11 | 0.01±0.002 | 0.37±0.06 | |||
| Standard length | 0.94±<0.0001 | 0.04 | Females | 0.38±0.20 | 35.5±19.6 | 65.9*±82.0* | 50.2±10.3 |
| Males | 0.42±0.24 | 36.5±21.7 | 64.6*±91.8* | 44.2±11.4 | |||
| Lateral plates | 0.99±<0.0001 | 0.01 | Females | 0.26±0.20 | 95.6±77.1 | 5.18*±3.7* | 0.02±0.004 |
| Males | 0.27±0.23 | 10.6±8.9 | 5.31*±4.3* | 0.02±0.005 |
Abbreviation: NA, not applicable.
Model did not converge for this estimate.
Values that significantly differ between the sexes are shown in bold. Note that the lengths were measured in centimetres.
Comparisons of G and P
Directions of maximum additive genetic variance were correlated between the sexes when all the body shape variables were considered (r=0.77, 95% confidence interval (CI): 0.16–0.97), although the corresponding angle between the directions of the vectors was different from zero (θ=37°, 95% CI: 15–80°). For the linear measurements, the correlation was lower (r=0.49, 95% CI: 0.024–0.92), and the angle between the directions deviated clearly from zero; θ=59° 95% CI: 23–89°). The correlation between sex-specific directions of maximum phenotypic divergence of the body shape variables (r=0.74, 95% CI: 0.05–0.98, θ=38°, 95% CI: 13–87°) was similar to that of directions of gmax. For the linear measurements, the correlation between directions of pmax vectors was higher (r=0.94, 95% CI: 0.14–0.99; θ=15° 95% CI: 6.2–82°). The difference between gmax and pmax was similar in both sexes (females: r=0.64, 95% CI: 0.04–0.97, θ=46°, 95% CI: 14–88° males: r=0.59, 95% CI: 0.04–0.96, θ=51°, 95% CI: 17–87°). The lack of significant differentiation between the G matrices and genetic correlations is also revealed by a heuristic inspection of the sex-specific matrices in Table 2. Similarly, the P matrix structures and phenotypic correlations did not differ significantly between the sexes (Table 3).
Table 2. Genetic variances (diagonal), covariances (upper off-diagonal) and correlations (lower off-diagonal) for the linear measurements in (a) females and (b) males.
| Anterior dorsal spine | Second dorsal spine | Head depth | Head length | Body depth | Jaw length | Pelvis length | Caudal peduncle | Dorsal fin | Snout length | |
|---|---|---|---|---|---|---|---|---|---|---|
| (a) | ||||||||||
| Anterior dorsal spine | 6.4 (1.31) | 5.0 (1.0) | 0.35 (0.57) | 0.21 (0.70) | 0.83 (0.79) | 0.38 (0.24) | 0.68 (0.49) | 1.7 (2.4) | 0.17 (1.19) | 0.53 (0.84) |
| Second dorsal spine | 0.92 (0.03) | 4.6 (0.93) | 0.14 (0.49) | 0.20 (0.59) | 0.55 (0.13) | 0.26 (0.41) | 0.67 (0.42) | 0.06 (0.00) | −0.05 (1.00) | 0.88 (0.71) |
| Head depth | 0.17 (0.27) | 0.08 (0.28) | 0.63 (0.13) | 0.34 (0.16) | 0.59 (0.19) | 0.62 (0.17) | 0.72 (0.32) | −0.85 (0.37) | 0.19 (0.37) | −0.19 (0.52) |
| Head length | 0.09 (0.28) | 0.10 (0.29) | 0.46 (0.17) | 0.84 (0.18) | 0.12 (0.27) | 0.26 (0.19) | 0.55 (0.38) | −0.76 | −0.35 (0.38) | 1.3 (0.47) |
| Body depth | 0.28 (0.26) | 0.23 (0.06) | 0.68 (0.16) | 0.12 (0.26) | 1.2 (0.23) | 0.66 (0.22) | 1.2 (0.42) | −1.1 (0.51) | 0.47 (0.25) | −0.19 (0.71) |
| Jaw length | 0.11 (0.07) | 0.09 (0.15) | 0.64 (0.10) | 0.23 (0.15) | 0.47 (0.12) | 1.6 (0.35) | 0.68 (0.54) | −0.86 (1.2) | 0.39 (0.15) | −0.40 (0.44) |
| Pelvis length | 0.21 (0.14) | 0.27 (0.15) | 0.72 (0.27) | 0.46 (0.32) | 0.90 (0.21) | 0.39 (0.30) | 1.7 (0.35) | −1.55 (0.66) | 1.1 (0.94) | 0.27 (0.87) |
| Caudal peduncle | 0.20 (0.28) | 0.01 (0.00)1 | −0.35 (0.13) | −0.26 (0.29) | −0.31 (0.13) | −0.20 (0.29) | −0.39 (0.13) | 10.4 (2.1) | 0.01 (0.00) | 0.71 (2.18) |
| Dorsal fin | 0.04 (0.27) | −0.01 (0.27) | 0.14 (0.28) | −0.23 (0.24) | 0.27 (0.13) | 0.20 (0.08) | 0.35 (0.29) | 0.002 (0.0003) | 2.9 (0.53) | −2.3 (0.90) |
| Snout length | 0.09 (0.15) | 0.18 (0.14) | −0.10 (0.27) | 0.60 (0.17) | −0.07 (0.27) | −0.14 (0.16) | 0.09 (0.29) | 0.09 (0.29) | −0.61 (0.21) | 5.1 (1.0) |
| (b) | ||||||||||
| Anterior dorsal spine | 6.8 (1.4) | 5.5 (1.16) | 0.94 (0.18) | 0.35 (0.83) | 0.79 (0.47) | 0.47 (1.14) | 1.9 (1.1) | 0.83 (2.80) | 0.87 (1.19) | −0.67 (1.02) |
| Second dorsal spine | 0.96 (0.02) | 5.0 (1.02) | 0.48 (0.29) | −0.08 (0.69) | 0.61 (0.00) | −0.63 (0.95) | 1.2 (0.91) | 1.9 (2.4) | 0.07 (0.99) | −0.57 (1.7) |
| Head depth | 0.34 (0.06) | 0.24 (0.14) | 1.1 (0.22) | 0.83 (0.23) | 1.0 (0.26) | 0.97 (0.12) | 0.96 (0.00) | −0.77 (1.04) | 0.08 (0.45) | −0.66 (0.77) |
| Head length | 0.12 (0.29) | −0.03 (0.29) | 0.75 (0.11) | 1.1 (0.25) | 1.1 (0.36) | 0.97 (0.35) | 0.68 (0.46) | −0.44 (1.10) | −0.46 (0.42) | 1.3 (0.78) |
| Body depth | 0.25 (0.14) | 0.23 (0.03) | 0.81 (0.14) | 0.86 (0.23) | 1.5 (0.29) | 1.2 (0.31) | 0.44 (0.27) | −1.02 (0.64) | −0.41 (0.53) | 0.07 (0.92) |
| Jaw length | 0.12 (0.28) | −0.18 (0.28) | 0.76 (0.06) | 0.61 (0.15) | 0.69 (0.09) | 2.3 (0.48) | 0.29 (0.64) | −0.80 (1.57) | −0.28 (0.70) | −0.61 (1.06) |
| Pelvis length | 0.73 (0.45) | 0.58 (0.43) | 0.97 (0.00) | 0.69 (0.51) | 0.36 (0.17) | 0.20 (0.44) | 1.2 (0.41) | −0.003 (0.00) | −0.47 (0.00) | 0.09 (0.00) |
| Caudal peduncle | 0.09 (0.31) | 0.25 (0.32) | −0.24 (0.30) | −0.13 (0.31) | −0.27 (0.15) | −0.16 (0.31) | −0.0012 (0.0003) | 10.1 (2.65) | −0.69 (1.45) | −0.94 (1.4) |
| Dorsal fin | 0.21 (0.28) | 0.02 (0.29) | 0.04 (0.26) | −0.28 (0.24) | −0.22 (0.28) | −0.12 (0.30) | −0.34 (0.00) | −0.13 (2.65) | 2.4 (0.48) | −2.9 (1.0) |
| Snout length | −0.10 (0.15) | −0.10 (0.30) | −0.22 (0.26) | 0.44 (0.20) | 0.02 (0.28) | −0.15 (0.26) | 0.03 (0.01) | −0.11 (0.17) | −0.73 (0.21) | 6.6 (1.4) |
Variances and covariances are × 103 their true value. Values significantly different from zero are shown in bold.
Table 3. Phenotypic variances (diagonal), covariances (upper off-diagonal) and correlations (lower off-diagonal) for the linear measurements in (a) females and (b) males.
| Anterior dorsal spine | Second dorsal spine | Head depth | Head length | Body depth | Jaw length | Pelvis length | Caudal peduncle | Dorsal fin | Snout length | |
|---|---|---|---|---|---|---|---|---|---|---|
| (a) | ||||||||||
| Anterior dorsal spine | 10.5 (0.71) | 6.9 (0.56) | 0.60 (0.17) | 0.19 (0.20) | 0.57 (0.22) | 0.40 (0.24) | 0.67 (0.27) | −1.09 (0.66) | 0.45 (0.32) | −0.05 (0.47) |
| Second dorsal spine | 0.76 (0.02) | 7.8 (0.51) | 0.41 (0.14) | 0.15 (0.17) | 0.57 (0.13) | 0.12 (0.24) | 0.84 (0.24) | 0.03 (0.00) | −0.07 (0.27) | 0.09 (0.40) |
| Head depth | 0.16 (0.04) | 0.13 (0.04) | 1.31 (0.08) | 1.1 (0.08) | 0.62 (0.07) | 0.90 (0.10) | −0.06 (0.09) | −0.90 (0.21) | 0.05 (0.10) | 0.74 (0.16) |
| Head length | 0.04 (0.04) | 0.04 (0.04) | 0.65 (0.02) | 2.1 (0.11) | 0.39 (0.08) | 0.84 (0.11) | −0.25 (0.11) | −0.85 (0.25) | −0.37 (0.13) | 2.4 (0.22) |
| Body depth | 0.14 (0.05) | 0.16 (0.04) | 0.43 (0.04) | 0.21 (0.04) | 1.6 (0.12) | 0.92 (0.12) | 0.55 (0.13) | −1.2 (0.28) | 0.28 (0.13) | 0.75 (0.20) |
| Jaw length | 0.06 (0.04) | 0.02 (0.04) | 0.40 (0.03) | 0.29 (0.04) | 0.36 (0.04) | 3.9 (0.22) | 0.03 (0.16) | −1.2 (0.37) | 0.01 (0.15) | 1.6 (0.27) |
| Pelvis length | 0.11 (0.04) | 0.16 (0.04) | −0.03 (0.04) | −0.09 (0.04) | 0.22 (0.04) | −.0.01 (0.04) | 3.6 (0.21) | −1.4 (0.38) | −0.001 (0.24) | −0.06 (0.26) |
| Caudal peduncle | −0.08 (0.05) | 0.003 (0.0001) | −0.18 (0.04) | −0.13 (0.04) | −0.21 (0.04) | −0.13 (0.04) | −0.16 (0.04) | 20.0 (1.2) | −4.1 (0.24) | −2.3 (0.62) |
| Dorsal fin | 0.07 (0.05) | −0.01 (0.05) | 0.02 (0.04) | −0.12 (0.04) | 0.11 (0.05) | 0.00 (0.04) | 0.00 (0.05) | 20.0 (0.02) | 4.2 (0.29 | −1.3 (0.32) |
| Snout length | −0.005 (0.04) | 0.01 (0.04) | 0.19 (0.04) | 0.49 (0.03) | 0.18 (0.05) | 0.25 (0.04) | −0.01 (0.04) | −0.15 (0.04) | −0.20 (0.04) | 11.0 (0.62) |
| (b) | ||||||||||
| Anterior dorsal spine | 10.2 (0.8) | 6.9 (0.63) | 0.93 (0.18) | 0.56 (0.24) | 0.78 (0.26) | 0.81 (0.33) | 0.5 (0.3) | −0.53 (0.77) | −0.05 (0.32) | −0.86 (0.56) |
| Second dorsal spine | 0.78 (0.02) | 7.8 (0.57) | 0.70 (0.17) | 0.57 (0.21) | 0.72 (0.10) | 0.68 (0.28) | 0.7 (0.28) | −0.7 (0.7) | −0.49 (0.27) | −0.21 (0.5) |
| Head depth | 0.23 (0.04) | 0.21 (0.05) | 1.6 (0.12) | 1.29 (0.12) | 0.8 (0.11) | 1.26 (0.12) | 0.09 (0.11) | −1.17 (0.31) | −0.23 (0.13) | 1.16 (0.24) |
| Head length | 0.12 (0.05) | 0.14 (0.05) | 0.69 (0.02) | 2.2 (0.15) | 0.5 (0.10) | 1.43 (0.17) | 0.22 (0.14) | −1.31 (0.34) | −0.60 (0.15) | 2.7 (0.30) |
| Body depth | 0.17 (0.05) | 0.19 (0.03) | 0.47 (0.04) | 0.26 (0.04) | 2.0 (0.16) | 1.2 (0.17) | 0.89 (0.15) | −2.10 (0.35) | 0.18 (0.14) | 0.95 (0.27) |
| Jaw length | 0.13 (0.05) | 0.12 (0.05) | 0.51 (0.03) | 0.48 (0.04) | 0.42 (0.04) | 4.0 (0.28) | 0.18 (0.19) | −1.99 (0.47) | −0.33 (0.19) | 2.36 (0.37) |
| Pelvis length | 0.07 (0.05) | 0.13 (0.05) | 0.03 (0.04) | 0.07 (0.04) | 0.31 (0.04) | 0.04 (0.05) | 4.2 (0.26) | −1.986 (0.28) | −0.67 (0.00) | 0.77 (0.20) |
| Caudal peduncle | −0.04 (0.05) | −0.05 (0.05) | −0.20 (0.05) | −0.19 (0.05) | −0.32 (0.05) | −0.21 (0.05) | −0.2078 (−0.2087) | 22.1 (1.53) | −3.72 (0.46) | −3.40 (0.8) |
| Dorsal fin | −0.01 (0.05) | −0.09 (0.05) | −0.09 (0.05) | −0.21 (0.05) | 0.07 (0.05) | −0.09 (0.05) | −0.17 (0.00) | −0.43 (0.04) | 3.6 (0.27) | −1.8 (0.4) |
| Snout length | −0.08 (0.05) | −0.02 (0.05) | 0.26 (0.05) | 0.52 (0.04) | 0.20 (0.05) | 0.34 (0.04) | 0.11 (0.03) | −0.21 (0.05) | −0.28 (0.05) | 11.7 (0.8) |
Variances and covariances are × 103 their true value. Values significantly different from zero are shown in bold.
Wild populations
SD in body shape in the wild populations was close to that observed in the lab (Figure 2). The main difference was observed in body depth, which did not differ as much in the wild as in the lab (Figures 2 and 3). The linear measurements confirmed the trend, with the exception of armour traits and standard length, which differed in the level of SD in different habitats and the lab. Generally, SD in armour traits was more pronounced in lakes and rivers as compared with the anadromic, sea and pond populations. In all populations, males were larger in traits related to head shape and size, while females had longer caudal peduncles and pelvic bones (Figure 3). In traits that were consistently larger in one sex across habitats (that is, head depth and length, snout length, jaw length, caudal peduncle length and pelvis length), SD was more pronounced in lake than pond populations (Figure 3). There was no significant correlation between the variance in SD of the linear measurements and lateral plates across the wild populations and rg(MF) of these traits measured in the laboratory population (rs=0.28, P=0.38).
Discussion
In theory, the evolution of SD is constrained by genomic conflict; the same genes code for the same traits in both sexes (Lande, 1980; Reeve and Fairbairn, 2001; Fairbairn et al., 2007). Although the general pattern seems to be that there is a negative correlation between the magnitude of the genetic correlation between the sexes and the level of SD (Poissant et al., 2010), there are some exceptions at the population level (Cowley et al., 1986; Cowley and Atchley, 1988; Ashman and Majetic, 2006), including this study. Also, positive correlation between rg(MF) and the level of SD was observed in 24 out of 66 studies by Poissant et al. (2010). Our results show that SD in morphological traits can evolve in natural populations despite strong genetic between-sex correlations. The present results also show that there is a heritable component to the SD despite strong inter-sex genetic correlations. In addition, the results indicate that the patterns of SD in wild populations cannot be predicted from the genetic correlations between the sexes in the ancestral population.
Patterns of SD in threespine stickleback
The general trends in the body shape and armour SD in the lab as well as in the wild populations of this study follow those reported in earlier studies of threespine sticklebacks from the wild (Caldecutt and Adams, 1998; Kitano et al., 2007; Aguirre et al., 2008; Spoljaric and Reimchen, 2008). Traits related to head shape and size are larger in males, while females have longer caudal peduncles and pelvises. The lengths of the dorsal spines are also larger in the lab-reared females compared with their male counterparts. Our results are also concordant with the only study that has measured sexual size dimorphism in the lab (Kitano et al., 2007). There was no sexual size dimorphism in the lab in either in this or Kitano et al. (2007) study, but it was observed in the wild, indicating that body size is more likely to be influenced by environmental factors than body shape.
The differences in the magnitude and direction of SD between habitats in armour traits are likely to reflect local differences in selection regimes. The anterior dorsal spine is usually shorter than the second dorsal spine, which forms a functional unit with the pelvic spines acting to protect the stickleback from gape-limited predators (Hoogland et al., 1957; Reimchen, 1983). In large lakes and marine habitats, the predator fauna is generally more complex than in ponds and rivers (reviewed by Reimchen, 1994), imposing stronger selection for the armour traits, which in turn is likely to constrain the evolution of SD in the second dorsal spine. At the same time, niche breadth is reduced in ponds compared with larger lakes, which in turn can act to reduce the extent of SD (Spoljaric and Reimchen, 2008). This trend is also reflected in our results—SD in body shape traits is more pronounced in lake populations compared with pond populations.
Although females that were clearly gravid with eggs were removed from the data set, condition and age are likely to affect the wild populations data set differently than the common garden fish, for which condition and age are standardized. This could cause discrepancies in the comparisons of females from the wild and from the laboratory populations. For instance, females from the wild might have deeper bodies because of past reproductive events. However, these types of (possible) condition-related differences are likely to be restricted to those aspects of body shape, which are affected by the volume of the abdominal cavity, and not influence traits connected to rigid bony structures, such as the length of the head or pelvic girdle.
The patterns of SD observed in the common garden fish are independent of condition and thus reflect genetically based differences between the sexes. SD of body depth therefore has genetic basis, which can be explained by the ecological differences between the sexes. Females are often more abundant in open water (for example, Reimchen, 1980), and thus exhibit a more limnetic lifestyle than males, which in turn have a more benthic lifestyle. Males should thus benefit from body shape (deep body, short and thick caudal peduncle, long snout, long jaw) more suitable for benthic lifestyle than females, whose body shape (shallow body, long and thin caudal peduncle, short snout, short jaw) reflects adaptation to limnetic habitats (for example, Walker, 1997). In fact, it has been suggested that the ancestral SD, as observed here in the common garden population, has provided variation on which selection could have acted on during the freshwater adaptation of threespine sticklebacks (Albert et al., 2008). Indeed, the patterns of SD in body shape closely follow those observed in between and within freshwater populations (for example, Walker, 1997; Leinonen et al., 2006; Spoljaric and Reimchen, 2008; Aguirre et al., 2008).
Contrary to the recent study on the genetic basis of SD in threespine stickleback body shape (Kitano et al., 2007), our results show that SD is present already before the onset of breeding. This suggests that SD in threespine sticklebacks is likely to be influenced by differential selection because of differences in ecology between the sexes, not only by sexual selection, as has been suggested for SD in threespine stickleback head shape (Caldecutt and Adams, 1998). Kitano et al. (2007) suggest that the occurrence of SD only after the onset of breeding implies that SD in body shape is a secondary sexual character possibly regulated by reproductive hormones. Our results are consistent with the idea that SD in traits related to body size requires the evolution of sex-specific patterns of growth and development and thus, SD in traits related to size are not restricted to the adult phase of the life cycle (Badyaev, 2002; Fairbairn et al., 2007).
Genetic constraints on the evolution of SD
Between-sex genetic correlations of the morphological traits measured here were high, with none of the rg(MF) estimates differing significantly from unity. This is congruent with the published estimates of rg(MF) for morphological traits, which are generally high (Roff, 1997; Lynch and Walsh, 1998; Poissant et al., 2010). The lack of significant association between the level of SD and rg(MF) can be explained by the high rg(MF) estimates or the relatively low absolute values of SD. For instance, although the meta-analysis by Poissant et al. (2010) showed a significant negative correlation between SD and rg(MF), the correlation appears to be get weaker, when only the highest rg(MF) estimates are considered (figure 4 in Poissant et al., 2010). The presence of SD despite strong rg(MF) has been thought to imply strong sex-specific selection leading to reduction in sex-specific genetic variation, which is followed by an increase in both SD and r g(MF) (Reeve and Fairbairn, 2001). This is a possible scenario also in the case of threespine sticklebacks. Differences in both sexual selection and natural selection (because of differences in ecology between sexes) have been suggested to contribute to the SD in threespine stickleback morphology (reviewed in Kitano et al., 2007; Spoljaric and Reimchen, 2008). Our results indicate that the role of differential selection in male and female threespine sticklebacks should be relatively large, as inferred from the evolution of SD in traits with high intersex genetic correlations. This conclusion is also supported by the similarities in male and female genetic variance–covariance matrices and directions of gmax, which in turn implies that if selection was similar in both sexes, they would be likely to exhibit similar, correlated responses to the selection.
A possible resolution to the genomic conflict in the threespine stickleback lies in the differential expression of genes, which has been shown to occur in an especially high frequency in the nascent sex chromosome (Leder et al., 2010), to which the majority of body shape and size QTL also mapped (Albert et al., 2008; Kitano et al., 2009). An example from cichlid fishes shows that sexual conflict can be resolved by evolution of a new sex determination locus (Roberts et al., 2009). It is thus possible that different levels of SD in the relatively recently derived (see Mäkinen et al., 2006) threespine stickleback populations of the present study reflect the relative ease with which the differences in the levels of SD can be achieved by regulating sex-limited expression (Albert et al., 2008). If this is the case, the strength of differential selection between the sexes would not have to be very strong, and SD could be achieved despite of the high intersex genetic correlations.
In conclusion, our results imply that SD of threespine stickleback body shape and armour can evolve despite of genomic conflict. Similarities in the genetic basis of body shape and armour traits in males and females do not appear to constrain the evolution of SD. It is likely that sex-specific selection pressures are strong enough to overcome the genetic constraints, and/or that sex-limited gene expression can aid evolution of SD in spite of the strong genetic correlations between the sexes. These explanations are not mutually exclusive, and a goal for future work lies in combining the study of the sex-limited gene expression and quantification of sex-specific selection. The genomic tools for doing this with threespine sticklebacks already exist, and are in use (for example, Peichel et al., 2004; Leder et al., 2009; Leder et al., 2010).
Acknowledgments
We thank Simo Rintakoski for processing the samples and help with practical issues. We are deeply indebted to Gábor Herczeg for helping in fish maintenance as well as insightful comments on earlier versions of this paper. We would also like to thank two anonymous reviewers for their valuable comments. Thanks are also due to Arthur Gilmour, Dean Adams, Patrick Phillips, Phillip Gienapp, Jussi Alho and Bob O'Hara for their assistance with statistical analyses and software. We also thank Chikako Matsuba for her assistance with the crosses and fish breeding, Marika Karjalainen for sexing the fish and John Loehr for checking the language. We are also grateful for the following people for collecting the stickleback samples: Linda Zaveik, Jörg Freyhof, Arne Levsen, Audrius Steponenas, Per-Arne Amundsen, Aleksei Veselov, Arne Nolte, Teija Aho, Ari Haikonen, Per Sjöstrand, Anders Berglund, Vladimír Kovács, Piotr Debowski, Dánjul Petur Højgaard, Volker Loeschcke, Ari Saura, Lennart Persson, Susie Coyle, Pat Monaghan, Dmitry Lajus and Thorsten Reusch. This study was supported by Academy of Finland (funding to JM), Ministry of Education (JM) and LUOVA Graduate School funded by the Ministry of Education (TL).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Aguirre WE, Ellis KE, Kusenda M, Bell MA. Phenotypic variation and sexual dimorphism in anadromous threespine stickleback: implications for postglacial adaptive radiation. Biol J Linn Soc. 2008;95:465–478. [Google Scholar]
- Albert AYK, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, et al. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution. 2008;62:76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- Anderson M. Sexual Selection. Princeton University Press: Princeton, NJ; 1994. [Google Scholar]
- Arnqvist G, Mårtensson T. Measurement error in geometric morphometrics: empirical strategies to assess and reduce its impact on measures of shape. Acta Zool Acad Sci Hung. 1998;44:73–96. [Google Scholar]
- Ashman TL. Constraints on the evolution of males and sexual dimorphism: field estimates of genetic architecture of reproductive traits in three populations of gynodioecious Fragaria virginiana. Evolution. 2003;57:2012–2025. doi: 10.1111/j.0014-3820.2003.tb00381.x. [DOI] [PubMed] [Google Scholar]
- Ashman TL, Majetic CJ. Genetic constraints on floral evolution: a review and evaluation of patterns. Heredity. 2006;96:343–352. doi: 10.1038/sj.hdy.6800815. [DOI] [PubMed] [Google Scholar]
- Badyaev A. Growing apart: an ontogenic perspective on the evolution of sexual size dimorphism. TREE. 2002;17:369–378. [Google Scholar]
- Bell MA. Lateral plate polymorphism and ontogeny of the complete plate morph of threespine sticklebacks (Gasterosteus aculeatus) Evolution. 1981;35:67–74. doi: 10.1111/j.1558-5646.1981.tb04859.x. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R, Rowe L. Intralocus sexual conflict and the genetic architecture of sexually dimorphic traits in Prochyliza xanthostoma (Diptera: Piophilidae) Evolution. 2005;59:1965–1975. [PubMed] [Google Scholar]
- Bookstein FL. Morphometric Tools for Landmark Data: Geometry and Biology. Cambridge University Press: Cambridge; 1991. [Google Scholar]
- Caldecutt WJ, Adams DC. Morphometrics of trophic osteology in the threespine stickleback, Gasterosteus aculeatus. Copeia. 1998;1998:827–828. [Google Scholar]
- Chapuis H, TixierBoichard M, Delabrosse Y, Ducrocq V. Multivariate restricted maximum likelihood estimation of genetic parameters for production traits in three selected turkey strains. Genet Sel Evol. 1996;28:299–317. [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Cowley DE, Atchley WR. Quantitative genetics of Drosophila melanogaster II. Heritabilities and genetic correlations between sexes for head and thorax traits. Genetics. 1988;119:421–433. doi: 10.1093/genetics/119.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley DE, Atchley WR, Rutledge JJ. Quantitative genetics of Drosophila melanogaster. 1. Sexual dimorphism in genetic parameters for wing traits. Genetics. 1986;114:549–566. doi: 10.1093/genetics/114.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, Amores A, Wilson C, Murphy J, Currey M, Phillips P, et al. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc Natl Acad Sci USA. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, Fuerst PA. Isolation of a sex-linked DNA sequence in cranes. J Hered. 2001;92:392–397. doi: 10.1093/jhered/92.5.392. [DOI] [PubMed] [Google Scholar]
- Fairbairn DJ.2007Sexual dimorphism in the water strider, Aquarius remigis: a case study of adaptation in response to sexually antagonistic selectionIn: Fairbairn DJ, Blanckenhorn WU, Szekely T (eds).Sex, Size and Gender Roles. Evolutionary Studies of Sexual Dimorphism Oxford University Press: Oxford [Google Scholar]
- Fairbairn DJ, Blanckenhorn WU, Szekely T.(eds) (2007Sex, Size and Gender Roles. Evolutionary Studies of Sexual Dimorphism Oxford University Press: Oxford [Google Scholar]
- Fairbairn DJ, Roff DA. The quantitative genetics of sexual dimorphism: assessing the importance of sex-linkage. Heredity. 2006;97:319–328. doi: 10.1038/sj.hdy.6800895. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Hermida M, Amaro R, San Miguel E. Lateral plate variation in Galician stickleback populations in the rivers Mino and Limia, NW Spain. Behaviour. 2000;137:965–979. [Google Scholar]
- Gibson JR, Chippindale AK, Rice WR. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc R Soc Lond Ser B Biol Sci. 2002;269:499–505. doi: 10.1098/rspb.2001.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour AR, Cullis BR, Welham SJ, Thompson R. ASREML Reference Manual. New South Wales Agriculture, Orange Agricultural Institute: Orange, NSW, Australia; 2001. [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R. ASReml User Guide Release 2.0. VSN International Ltd: Hemel Hempstead, HP1 1ES, UK; 2006. [Google Scholar]
- Greenwood PJ, Adams J. Sexual Selection, size dimorphism and a fallacy. Oikos. 1987;48:106–108. [Google Scholar]
- Hagen DW. Inheritance of numbers of lateral plates and gill rakers in Gasterosteus aculeatus. Heredity. 1973;30:303–312. [Google Scholar]
- Hedrick AV, Temeles EJ. The evolution of sexual dimorphism in animals—hypotheses and tests. Trends Ecol Evol. 1989;4:136–138. doi: 10.1016/0169-5347(89)90212-7. [DOI] [PubMed] [Google Scholar]
- Hoogland R, Morris D, Tinbergen N. The spines of sticklebacks (Gasterosteus and Pygosteus) as a means of defense against predators (Perca and Esox) Behaviour. 1957;10:205–236. [Google Scholar]
- Kitano J, Mori S, Peichel CL. Sexual dimorphism in the external morphology of the threespine stickleback (Gasterosteus aculeatus) Copeia. 2007;2007:336–349. [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP, Leamy LJ. Quantitative genetics of geometric shape in the mouse mandible. Evolution. 2001;55:2342–2352. doi: 10.1111/j.0014-3820.2001.tb00747.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, McIntyre GS. Geometric morphometrics of developmental instability: analyzing patterns of fluctuating asymmetry with Procrustes methods. Evolution. 1998;52:1363–1375. doi: 10.1111/j.1558-5646.1998.tb02018.x. [DOI] [PubMed] [Google Scholar]
- Lande R. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution. 1980;34:292–305. doi: 10.1111/j.1558-5646.1980.tb04817.x. [DOI] [PubMed] [Google Scholar]
- Leder E, Merilisä J, Primmer C. A flexible whole-genome microarray for transcriptomics in three-spine stickleback (Gasterosteus aculeatus) BMC Genomics. 2009;10:426. doi: 10.1186/1471-2164-10-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder EH, Cano JM, Leinonen T, O'Hara RB, Nikinmaa M, Primmer CR, et al. Biased expression on the X chromosome as a key step in sex chromosome evolution. Mol Biol Evol. 2010;27:1495–1503. doi: 10.1093/molbev/msq031. [DOI] [PubMed] [Google Scholar]
- Leinonen T, Cano JM, Mäkinen HS, Merilä J. Contrasting patterns of body shape and neutral genetic divergence in marine and lake populations of threespine sticklebacks. J Evol Biol. 2006;19:1803–1812. doi: 10.1111/j.1420-9101.2006.01182.x. [DOI] [PubMed] [Google Scholar]
- Lovich JE, Gibbons JW. A review of techniques for quantifying sexual size dimorphism. Growth Dev Aging. 1992;56:269–281. [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates: Sunderland, MA; 1998. [Google Scholar]
- Mäkinen HS, Cano JM, Merilä J. Genetic relationships among marine and freshwater populations of the European three-spined stickleback (Gasterosteus aculeatus) revealed by microsatellites. Mol Ecol. 2006;15:1519–1534. doi: 10.1111/j.1365-294X.2006.02871.x. [DOI] [PubMed] [Google Scholar]
- Mäkinen HS, Merilä J. Mitochondrial DNA phylogeography of the three-spined stickleback (Gasterosteus aculeatus) in Europe - evidence for multiple glacial refugia. Mol Phylogenet Evol. 2008;46:167–182. doi: 10.1016/j.ympev.2007.06.011. [DOI] [PubMed] [Google Scholar]
- McDaniel SF. Genetic correlations do not constrain the evolution of sexual dimorphism in the moss Ceratodon purpureus. Evolution. 2005;59:2353–2361. [PubMed] [Google Scholar]
- McGuigan K, Chenoweth SF, Blows MW. Phenotypic divergence along lines of genetic variance. Am Nat. 2005;165:32–43. doi: 10.1086/426600. [DOI] [PubMed] [Google Scholar]
- Mealey L. Sex differences. Developmental and Evolutionary Strategies. Academic Press: San Diego, CA; 2000. [Google Scholar]
- Moodie GEE. Predation, natural selection and adaptation in an unusual threespine stickleback. Heredity. 1972;28:155–167. [Google Scholar]
- Moodie GEE, Reimchen TE. Glacial refugia, endemism, and stickleback populations of Queen Charlotte Islands, British Columbia. Can Field Nat. 1976;90:471–474. [Google Scholar]
- Münzing J. Evolution of variation and distributional patterns in European populations of three-spined stickleback, Gasterosteus aculeatus. Evolution. 1963;17:320–332. [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, et al. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, et al. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Pimentel RA. Morphometrics. Kendall/Hunt Publishing Co: Dubuque; 1979. [Google Scholar]
- Poissant J, Wilson AJ, Coltman DW. Sex-specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross-sex genetic correlations. Evolution. 2010;64:97–107. doi: 10.1111/j.1558-5646.2009.00793.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JR, Schluter D. Declining interspecific competition during character displacement: summoning the ghost of competition past. Evol Ecol Res. 2001;3:209–220. [Google Scholar]
- Reeve JP, Fairbairn DJ. Predicting the evolution of sexual size dimorphism. J Evol Biol. 2001;14:244–254. [Google Scholar]
- Reimchen TE. Spine deficiency and polymorphism in a population of Gasterosteus aculeatus—an adaptation to predators. Can J Zool. 1980;58:1232–1244. [Google Scholar]
- Reimchen TE. Structural relationships between spines and lateral plates in threespine stickleback (Gasterosteus aculeatus) Evolution. 1983;37:931–946. doi: 10.1111/j.1558-5646.1983.tb05622.x. [DOI] [PubMed] [Google Scholar]
- Reimchen TE.1994Predators and evolution in threespine sticklebackIn: Bell MA, Foster SA (eds).The Evolutionary Biology of Threespine Stickleback Oxford University Press: Oxford [Google Scholar]
- Reimchen TE, Nosil P. Replicated ecological landscapes and the evolution of morphological diversity among Gasterosteus populations from an archipelago on the west coast of Canada. Can J Zool. 2006;84:643–654. [Google Scholar]
- Reimchen TE, Stinson EM, Nelson JS. Multivariate differentiation of parapatric and allopatric populations of threespine stickleback in the Sangan River Watershed, Queen Charlotte Islands. Can J Zool. 1985;63:2944–2951. [Google Scholar]
- Reusch TBH, Wegner KM, Kalbe M. Rapid genetic divergence in postglacial populations of threespine stickleback (Gasterosteus aculeatus): the role of habitat type, drainage and geographical proximity. Mol Ecol. 2001;10:2435–2445. doi: 10.1046/j.0962-1083.2001.01366.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff DA. Evolutionary Quantitative Genetics. Chapman and Hall: New York; 1997. [Google Scholar]
- Rohlf FJ. TPS Software Series. Department of Ecology and Evolution, State University of New York, Stony Brook; 2006. [Google Scholar]
- Rohlf FJ. TPS Software Series. Department of Ecology and Evolution, State University of New York: Stony Brook, NY; 2007. [Google Scholar]
- Rohlf FJ, Slice D. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst Zool. 1990;39:40–59. [Google Scholar]
- Schluter D. Adaptive radiation along genetic lines of least resistance. Evolution. 1996;50:1766–1774. doi: 10.1111/j.1558-5646.1996.tb03563.x. [DOI] [PubMed] [Google Scholar]
- Schluter D, Clifford EA, Nemethy M, McKinnon JS. Parallel evolution and inheritance of quantitative traits. Am Nat. 2004;163:809–822. doi: 10.1086/383621. [DOI] [PubMed] [Google Scholar]
- Sharpe DMT, Räsänen K, Berner D, Hendry AP. Genetic and environmental contributions to the morphology of lake and stream stickleback: implications for gene flow and reproductive isolation. Evol Ecol Res. 2008;10:849–866. [Google Scholar]
- Short RV, Balabban E. The Differences Between the Sexes. Cambridge University Press: Cambridge; 1994. [Google Scholar]
- Slatkin M. Ecological causes of sexual dimorphism. Evolution. 1984;38:622–630. doi: 10.1111/j.1558-5646.1984.tb00327.x. [DOI] [PubMed] [Google Scholar]
- Spoljaric MA, Reimchen TE. Habitat-dependent reduction of sexual dimorphism in geometric body shape of Haida Gwaii threespine stickleback. Biol J Linn Soc. 2008;95:505–516. [Google Scholar]
- Valentin AE, Penin X, Chanut JP, Sevigny J-P, Rohlf FJ. Arching effect on fish body shape in geometric morphometric studies. J Fish Biol. 2008;73:623–638. [Google Scholar]
- Walker JA. Ecological morphology of lacustrine threespine stickleback Gasterosteus aculeatus L (Gasterosteidae) body shape. Biol J Linn Soc. 1997;61:3–50. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.