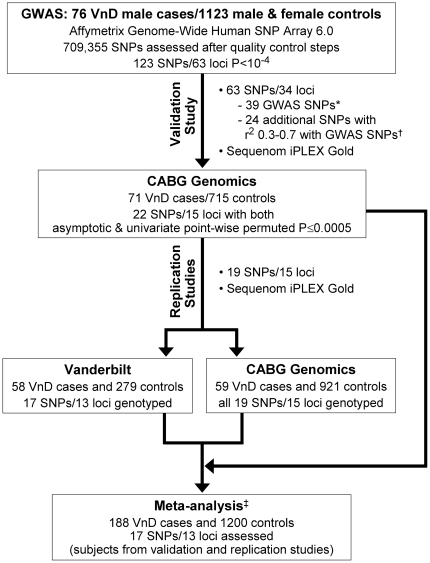
Figure 1. Schematic illustrating sequence of GWAS study followed by validation study, replication studies, and meta-analysis.
All subjects were of European ancestry and were male unless otherwise specified. * 39 Affymetrix 6.0 SNPs associated with VnD in the GWAS (P<10−4) or SNPs in strong linkage disequilibrium (r2>0.80; estimated using HaploView 4.1.) with these GWAS SNPs were genotyped from 34 genetic loci for further validation study assessment. † In order to identify SNPs with potentially stronger associations with VnD within some of the genetic loci identified in the GWAS, additional SNPs were genotyped that were in moderate linkage disequilibrium with associated GWAS SNPs (r2 = 0.30–0.70). Correlations (r2) were estimated using HaploView 4.1. ‡ SNP associations with VnD were analyzed by enrolling institution (BWH, THI, and Vanderbilt University Medical Center) with adjustments for age, preoperative LVEF, and duration of CPB time, and then combined by meta-analysis.