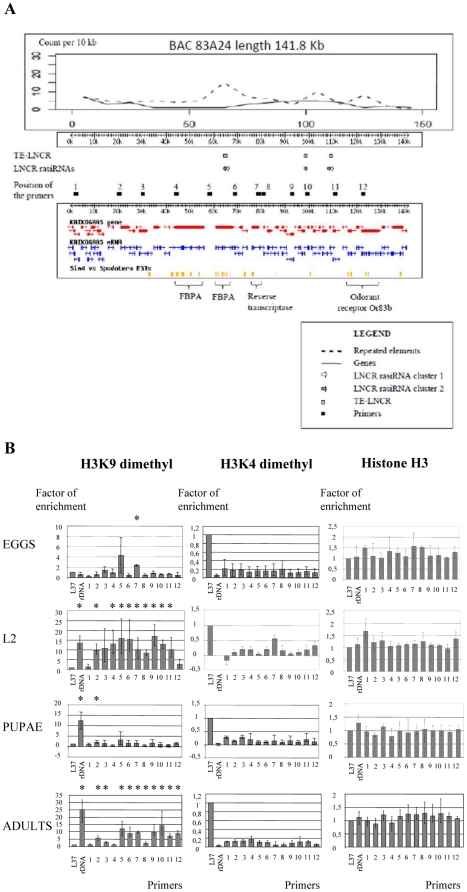
Figure 5. Distribution of genes, repeated elements, euchromatic and heterochromatic markers in genomic region cloned in BAC 83A24.
A) Schematic presentation of genetic and epigenetic elements in the genomic region cloned in BAC 83A24 (141.8 Kb). Upper panel presents distribution of genes and DNA repeated elements in genomic region cloned in BAC 83A24. The positions of TE-LNCR, LNCR rasiRNAs and primers used in ChIP qPCR are indicated in the middle. The lower panel of the figure presents this genomic region with the position of predicted genes and mRNAs, as well as the position of S. frugiperda ESTs (taken from LepidoDB (http://genouest.org/cgi-bin/gbrowse/gbrowse/lepido_pub/)). The positions of putative genes are indicated below (FBPA gene is duplicated). B) Presence of histone H3 and histone H3 modification (H3K9me2 and H3K4me2) in analyzed genomic regions (BAC 83A24) during different developmental stages done by ChIP-qPCR. L37 ribosomal gene was used as a positive control for euchromatic regions and rDNA as a positive control of heterochromatic regions. Error bars indicate standard deviations over at least three independent experiments. Asterisks indicate statistically significant enrichment of heterochromatic marker over the control region L37 (p<0,05). P-values were calculated using Student's t-test.