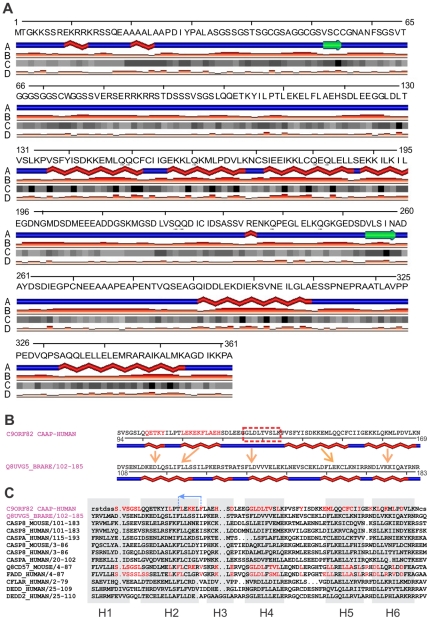
Figure 2. Overall structure of CAAP.
(A) SABLE (http://sable.cchmc.org) was used to predict the structure of CAAP. The amino acid sequence, predicted secondary structures (with helices shown as red braids, beta strands as green arrows, and loops as blue lines) (row A), the confidence of secondary structure predictions at each position (row B, the higher the bar the more confident the prediction), predicted solvent accessibilities (row C, black boxes corresponding to fully buried positions, and shades of grey representing the degree of solvent exposure), and the confidence bars for the solvent accessibility prediction (row D, the higher the bar the more confident the prediction), are shown in the respective rows in the figure. Alignment of the central CAAP domain. (B) FFAS alignment of the central CAAP domain and a DED family representative Q8UVG5 (also indicated in blue in Fig. 2C). The SABLE and PsiPRED (http://bioinf.cs.ucl.ac.uk/psipred/) predicted helices (the latter highlighted using red font for amino acid residues) within the central CAAP domain are shown as red braids. As can be seen from the Figure, they are qualitatively consistent with those of the DED matching domain shown for comparison below. However, due to low homology, FFAS likely misaligns some of the helical regions, as indicated by arrows between predicted and observed helices (the latter also shown in the context of multiple alignment in Fig. 2C); Alignment of the DED domains (C) Multiple alignment of DED domains from several members of the DD superfamily, superimposed with the approximate position of canonical six helices of DD [2], which are shown as gray boxes. Note that CAAP sequence, which is shown on top (using FFAS alignment into Q8UVG5), is consistent with the range of substitutions observed within the family, and also that it appears to be most consistent with its FADD members (human and mouse), as indicated in red to highlight several conserved motifs. Note also, that several functionally relevant motifs, such as FLAEH (residues 115–119) or KxL (residues 157–159), appear to be misaligned in the automatically generated FFAS alignment.