SUMMARY
It has been widely assumed that all transcription in cells occurs using NTPs only, i.e. de novo. However, it has been known for several decades that both prokaryotic and eukaryotic RNA polymerases (RNAPs) can utilize small (2- to ~5-nt) RNAs to prime transcription initiation in vitro, raising the possibility that small RNAs might also prime transcription initiation in vivo. A new study by Goldman et al. has now provided the first evidence that priming with so-called “nanoRNAs”, i.e. 2- to ~5-nt RNAs, can, in fact, occur in vivo. Furthermore, this study provides evidence that altering the extent of nanoRNA-mediated priming of transcription initiation can profoundly influence global gene expression. In this perspective we summarize the findings of Goldman et al. and discuss the prospect that nanoRNA-mediated priming of transcription initiation represents an under appreciated aspect of gene expression in vivo.
Introduction: a historical perspective on priming of transcription with small RNAs
Multisubunit DNA-dependent RNA polymerases (RNAPs) were first identified in 1960 based on their ability to synthesize RNA transcripts in vitro in the presence of a DNA template and NTPs 1. In addition to being able to initiate RNA synthesis using NTPs only, i.e. “de novo”, it was demonstrated in the mid-1960s that RNAP could also initiate transcription by using a 2- to 5-nt RNA oligonucleotide carrying a 3′ hydroxyl group as a primer 2, 3. Later studies revealed that all multisubunit DNA-dependent RNAPs--bacterial RNAP, eukaryotic RNAP I, RNAP II, RNAP III, and archael RNAP--can use small RNA oligonucleotides to prime promoter-specific transcription initiation in vitro 4–7. Furthermore, the use of such small RNAs to prime transcription initiation in vitro has become commonplace for researchers studying transcription.
In the mid-1970s it was proposed that priming of transcription with small RNAs occurs in cells and can influence gene expression 8, 9. In particular, it was proposed that small RNA fragments derived from the processing of longer transcripts could serve as sequence-specific primers that stimulate transcription initiation from specific target promoters 8, 9. The proposal that small RNA fragments could “activate” transcription in this manner was based upon the empirical observation that yields of RNA transcripts produced in vitro could, in certain cases, be increased by adding a small RNA primer to the transcription reactions 2, 3, 10, 11. Nevertheless, in the ~35 years since this proposal was made, no evidence that small RNAs can actually prime transcription initiation in vivo has been presented. Thus, the generally accepted view has been that all transcription in cells occurs de novo.
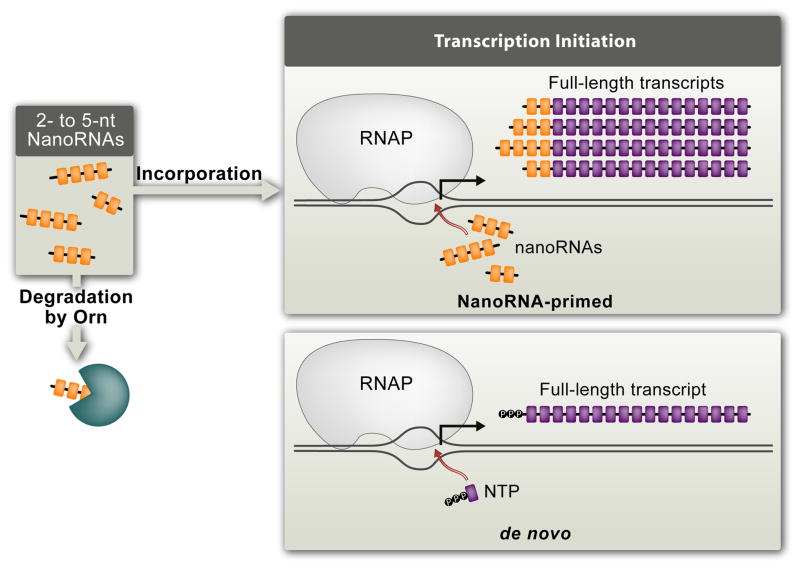
A new study by Goldman et al. has now challenged the conventional paradigm that all cellular transcription occurs de novo 12. In particular, this study has provided evidence that very small RNAs can, in fact, prime transcription initiation in vivo. The findings of Goldman et al. also provide evidence that altering the extent to which transcription initiation is primed or occurs de novo can significantly impact gene expression. These new findings provide an impetus to revisit the notion that priming of transcription with very small RNAs might be an important contributor to cellular gene expression.
Evidence that nanoRNAs prime transcription initiation in vivo
To demonstrate that very small RNAs can be used by RNAP as primers for transcription initiation in vivo, Goldman et al. took advantage of studies investigating the metabolism of small RNAs in bacterial cells. These studies had revealed the existence of a class of enzymes responsible for the degradation of RNAs between 2 and ~5-nt in length 13–17; such extremely small RNAs have been termed “nanoRNAs” to distinguish them from other classes of small RNAs 16. In E. coli the degradation of nanoRNAs is carried out by an essential 3′ to 5′ exonuclease, Oligoribonuclease (Orn) 15. Thus, inactivation of Orn in E. coli was shown to result in the accumulation of 2- to ~5-nt nanoRNAs 15. Accordingly, Goldman et al. tested whether inactivation of Orn might lead to cellular conditions where a significant fraction of transcription is primed due to the presence of elevated concentrations of 2- to ~5-nt nanoRNAs. Because inactivation of Orn in E. coli leads to a loss of viability 15, the authors determined the effect of inactivating Orn in Pseudomonas aeruginosa, a Gram-negative organism that remains viable in the absence of Orn function 18.
Using a specially engineered strain of P. aeruginosa that allows for the artificial depletion of Orn, Goldman et al. first demonstrated that, similar to results obtained in E. coli, loss of Orn function in P. aeruginosa leads to accumulation of 2- to ~4-nt nanoRNAs. Next, to determine whether increasing the intracellular concentrations of 2-to ~4-nt nanoRNAs leads to the widespread use of nanoRNAs as primers for transcription initiation, the authors used high-throughput sequencing to analyze the 5′ ends of primary transcripts on a genome-wide scale.
In vitro studies indicate that 2- to 4-nt RNAs that are complementary to the DNA template can efficiently compete with NTPs for use as primers during transcription initiation provided the 5′ end of the RNA base pairs with promoter sequences between positions −3 and +1 and the 3′ end of the RNA base pairs with promoter sequences between positions +1 and +3 11, 12, 19–23. (Note that by convention the position where de novo transcription begins is designated +1.) Thus, use of 2- to 4-nt RNAs to prime transcription initiation in vitro can, in some cases, result in production of transcripts that are extended in length by 1, 2, or 3 nt at the 5′ end compared with transcripts that are produced in reactions performed in the presence of NTPs alone. The authors therefore reasoned that if 2- to 4-nt RNAs could also prime transcription initiation in vivo, increasing the intracellular concentration of 2- to 4-nt nanoRNAs should result in 5′ end alterations for primary transcripts produced from a significant fraction of cellular promoters. Consistent with this prediction, using high-throughput sequencing Goldman et al. found that depletion of Orn did, in fact, cause the appearance of significant amounts of primary transcripts that were, on average, 1, 2, or 3 nt longer than those observed in non-depleted cells.
The authors presented several lines of evidence supporting the conclusion that the observed changes in the average distributions of the 5′ ends of primary transcripts upon depletion of Orn are a direct consequence of widespread use of 2- to 4-nt nanoRNAs as primers for transcription initiation. Among these, the two most important are the following. First, depleting Orn in the presence of a heterologous nanoRNA-degrading enzyme, the NrnB protein from Bacillus subtilis 17, did not result in global alterations in the average distributions of the 5′ ends of primary transcripts. This finding established that the observed change in the distribution of the 5′ ends of transcripts that occurs upon depletion of Orn was a direct consequence of the accumulation of 2- to ~4-nt nanoRNAs (and not due to an unrelated consequence of Orn depletion). Second, it is well documented that alterations in the intracellular concentrations of NTPs can lead to changes in the position where transcription initiation begins [reviewed in 24]. (This phenomenon, which is termed NTP-dependent start point switching, is discussed further below.) Thus, it was important to rule out the possibility that the observed changes in the average distributions of the 5′ ends of primary transcripts upon depletion of Orn were not due simply to any alterations in NTP concentrations. To investigate this possibility, Goldman et al. compared the average distribution of the 5′ ends of transcripts carrying a 5′ triphosphate group, i.e. those potentially derived from a de novo initiation event, with the average distribution of the 5′ ends of all transcripts regardless of the phosphorylation status of the 5′ end. Strikingly, this comparison revealed that whereas depletion of Orn results in a pronounced alteration in the distribution of the 5′ ends of transcripts when all 5′ ends are considered, depletion of Orn results in only a modest change in the average distribution of transcripts carrying a 5′ triphosphate group. These data rule out the possibility that the pronounced changes in the average distributions of the 5′ ends of transcripts are a consequence of alterations in the abundance of NTPs that, in turn, alters the position of de novo initiation. Consistent with this assertion, the authors presented evidence that the intracellular concentrations of NTPs were identical in Orn-depleted cells and cells in which Orn was not depleted.
NanoRNA-mediated priming as a means of altering gene expression
Having shown that depletion of Orn leads to widespread nanoRNA-mediated priming, Goldman et al. next determined the effect that widespread nanoRNA-mediated priming had on global gene expression. Using DNA microarrays they found that the relative abundance of transcripts associated with ~20% of all genes differed by a factor of 2 of more when comparing Orn-depleted cells with non-depleted cells. Importantly, nearly all of the genes whose expression changed significantly upon depletion of Orn did not exhibit a significant change in expression when Orn was depleted in the presence of a heterologous nanoRNA-degrading enzyme. Thus, the authors concluded that the vast majority of the changes in gene expression that occur upon depletion of Orn are a direct consequence of the accumulation of 2- to ~4-nt nanoRNAs. The expression analysis therefore revealed a correlation between widespread nanoRNA-mediated priming of transcription initiation and widespread alterations in gene expression; these findings raise the intriguing possibility that the balance between de novo initiation and nanoRNA-primed initiation may be an important determinant of gene expression.
Below we consider several potential mechanisms by which altering the extent of nanoRNA-primed transcription at a particular promoter could alter gene expression (Figure 1).
Figure 1.
NanoRNA-mediated priming of transcription initiation alters gene expression. NanoRNAs (2- to 5-nt RNAs) can either be degraded by Oligoribonuclease (Orn) or used by RNAP as primers to initiate transcription. Compared with initiation with NTPs only (i.e. de novo), initiation with nanoRNAs could, in principle, alter gene expression by increasing yields of transcripts, altering the sequence of the 5′ ends of transcripts, or altering the phosphorylation status of the 5′ ends of transcripts.
NanoRNA-mediated priming as a means of altering transcription efficiency
As mentioned above, ~35 years ago the proposal that small RNA oligonucleotides might “activate” transcription from specific target promoters was made. In support of this proposal biochemical evidence indicates that the affinity of nanoRNAs for the RNAP-promoter open complex can be substantially higher than that of the initiating mononucleotide. In particular, the affinity of a 2-nt nanoRNA complementary to positions +1 and +2 for an RNAP-promoter open complex can be ~10-fold higher than the affinity of a mononucleotide complementary to position +1; furthermore, the affinity of a 3-nt nanoRNA complementary to positions +1, +2 and +3 for a RNAP-promoter open complex can be ~100-fold higher than the affinity of a mononucleotide complementary to position +1 23, 25. Correspondingly, the half-maximal effective concentration for initiating transcription (Km) of a 2-nt nanoRNA complementary to positions +1 and +2, or a 3-nt nanoRNA complementary to positions +1, +2, and +3, can be ~10- to 100-fold lower than the Km of a mononucleotide complementary to position +1. A regulatory mechanism that involves nanoRNAs acting to stimulate transcription initiation may be targeted to promoters that require high concentrations of the initiating NTP (and/or the second NTP added to the transcript) for efficient de novo initiation.
We note that, in contrast to nanoRNAs that carry a 3′ hydroxyl group, those that carry either a 3′ deoxy or 3′ phosphate group will not be able to function as a primers for transcription initiation. Such “non-extendable” nanoRNA species could, in principle, function as promoter-specific repressors. In this regard, it has been shown that 3′ deoxy small RNA oligonucleotides complementary to the region around the transcription start point can be potent promoter-specific inhibitors of transcription in vitro 26, 27. In this case, inhibition of transcription occurs due to the hybridization of the non-extendable oligonucleotide with the exposed single-stranded DNA that is generated during formation of the RNAP-promoter open complex, which traps the transcription complex in an inactive state.
NanoRNA-mediated priming as a means of altering the sequence of the 5′ end of a transcript
As illustrated in the study by Goldman et al., initiation with a nanoRNA primer provides a means by which the sequence of the 5′ end of the transcript can be altered. Specifically, Goldman et al. found that increasing the extent of nanoRNA-mediated priming leads to the production of transcripts that are 1, 2, or 3 nucleotides longer than their de novo initiated counterparts. In principle, the nanoRNA-mediated addition of 1, 2, or 3 nucleotides onto the 5′ end of a transcript could have a significant impact on gene expression by a variety of mechanisms. For example, the addition of nucleotides onto the 5′ end of a transcript could profoundly influence the secondary structure of the resultant RNA transcript and thereby alter transcript stability. In this regard, unpaired 5′ ends can strongly destabilize transcripts in E. coli by increasing the likelihood that the transcript will be cleaved by RNase E, the primary endonuclease in this organism 28, 29. The nanoRNA-mediated addition of nucleotides onto the 5′ end of a transcript could also influence events that occur during transcription elongation. Indeed, it is well established that changes in the sequence of the 5′ end of a transcript can inhibit the efficiency of an intrinsic terminator located several hundred base pairs downstream 30, 31. In these cases it is likely that the 5′ end of a transcript, due to its inherent flexibility, is able to hybridize with other regions of the RNA, which, in turn, prevents formation of the terminator hairpin.
Precedent for the model that the nanoRNA-mediated addition of 1, 2, or 3 nucleotides onto the 5′ end of a transcript could influence gene expression also comes from studies of NTP-dependent transcription start-point switching (reviewed in 24). In particular, the two examples of NTP-dependent transcription start-point switching provided below reveal how the addition of 1 or 2 nt onto the 5′ end of a transcript can significantly influence gene expression by altering either translation efficiency or the extent to which productive transcription occurs.
First, we consider the case of the pyrC promoters of E. coli and Salmonella enterica, which control expression of a gene involved in pyrimidine biosynthesis. Production of the pyrC gene product is determined by the intracellular concentration of CTP, which dictates whether transcription from the pyrC promoter will initiate at one of two template positions that are separated by 2 base pairs (bp) 32, 33. When CTP concentrations are high transcription initiates from a C located 7 bp downstream of the promoter 10 element (C7), whereas when CTP concentrations are low transcription initiates from a G located 9 bp downstream of the promoter −10 element (G9). Primary transcripts initiating from C7 are not efficiently translated due to formation of an RNA secondary structure that inhibits ribosome binding. In contrast, transcripts initiated from position G9 are efficiently translated because these transcripts cannot adopt a secondary structure that inhibits ribosome binding. Thus, in the case of the pyrC promoter, the addition of 2 nt to the 5′ end of the RNA transcript dramatically alters protein synthesis.
Second, we consider the case of the codB promoter of E. coli, which controls the expression of genes involved in cytosine uptake and utilization. Gene expression from the codB promoter is determined by the intracellular concentration of UTP, which determines whether transcription will initiate at one of two template positions that are separated by 1 bp 34. In particular, when the concentration of UTP is sufficiently high, transcription initiates from an A located 8 bp downstream of the promoter 10 element (A8); this A is followed by a stretch of 6 Ts. (Note that in this case UTP is the second nucleotide added to the nascent RNA transcript and is not the initiating nucleotide.) Initiation at A8 leads to non-productive transcription through a mechanism of UTP-dependent reiterative transcription. Specifically, transcription of the 6 T:A base pairs in the initial transcribed region of codB creates a weak RNA-DNA hybrid that is prone to ‘slippage’. As a result of transcript slippage at this poly T tract, UTP is repetitively added to the 3′ end of the nascent RNA transcript, ultimately resulting in the release of the transcript. In contrast, when the UTP concentration is relatively low, transcription initiates from a G located 1 bp upstream of A8 and 7 bp downstream of the promoter 10 element (G7). When transcription initiates at G7, UTP-dependent reiterative transcription does not occur and the result is production of full-length transcripts. Initiation at G7 is thought to prevent reiterative transcription due to the addition of a stabilizing G:C base pair in the RNA-DNA hybrid, which anchors the transcript and prevents slippage upon the encounter of the poly-T stretch. Thus, in the case of the codB promoter, the addition of a single nucleotide to the 5′ end of the RNA transcript enables productive transcription to occur.
NanoRNA-mediated priming as a means of altering the phosphorylation state of the transcript 5′ end
Goldman et al. observed pronounced alterations in the distribution of transcript 5′ ends when all transcripts were analyzed regardless of the phosphorylation status of the 5′ end and not when only transcripts carrying a 5′ triphosphate group were analyzed. These results indicate that the majority of nanoRNAs that prime transcription upon depletion of Orn carry either a 5′ monophosphate or a 5′ hydroxyl group. Furthermore, these results suggest that widespread nanoRNA-mediated priming of transcription results in the production of a significant fraction of primary transcripts that do not carry a 5′ triphosphate.
The phosphorylation state of the 5′ end of primary transcripts is a key determinant of transcript stability [for review see 35]. In particular, the rate-limiting step in the degradation of many mRNA transcripts in E. coli is the conversion of the 5′ end of the transcript from a triphosphate to a monophosphate, which stimulates the activity of RNase E 29, 36. In E. coli, conversion of the 5′ end of the transcript from a triphosphate to a monophosphate can occur through the action of the RNA pyrophosphohydrolase RppH 29. Thus, in principle, incorporation of a 5′ monophosphate-carrying nanoRNA could provide a means of bypassing the requirement for RppH and accelerating RNA degradation. In contrast, as RNase E does not efficiently cleave RNA transcripts carrying a 5′ hydroxyl end 36–38, incorporation of a 5′ hydroxyl-carrying nanoRNA could provide a means of stabilizing the resultant RNA transcript.
We note that in vitro analysis indicates that incorporation of NMPs or nucleosides into the 5′ end of the nascent transcript is strongly disfavored. Thus the Km of an NTP that is complementary to position +1 is, in general, an order of magnitude lower than the Km for the corresponding NMP or nucleoside 39. In contrast, nanoRNAs (of identical sequence) that carry a 5′ triphosphate, a 5′ monophosphate or a 5′ hydroxyl group are equally efficient as primers for transcription initiation 23. Thus, whereas de novo initiation events are biased against the incorporation of a 5′ monophosphate or a 5′ hydroxyl group into the resultant RNA transcript, nanoRNA-mediated priming events would not exhibit such a bias. Accordingly, nanoRNA-mediated priming provides a mechanism by which the phosphorylation state of the transcript 5′ end could be readily altered. Furthermore, an initiation event that utilizes a 2-nt nanoRNA complementary to positions +1 and +2, or a 3-nt RNA oligo complementary to positions +1, +2, and +3 could enable the phosphorylation state of the 5′ end of a transcript to change without any alteration in the sequence of the 5′ end.
Potential sources of nanoRNAs
In principle, there are several cellular processes that could generate nanoRNA species carrying a 3′ hydroxyl group that would be suitable for use as a primer.
Intermediates of RNA degradation
As mentioned above, it has previously been proposed that small RNA fragments derived from the processing of longer transcripts could serve as sequence-specific primers that stimulate transcription initiation from specific target promoters. In this regard, nanoRNAs are, in fact, likely generated in vivo as intermediates during mRNA degradation. In particular, the canonical pathway for the degradation of mRNA transcripts in E. coli begins with the endonucleolytic cleavage of a full-length transcript by RNase E, which cleaves the full-length transcript several times in single-stranded regions to generate a number of smaller fragments [reviewed in 35, 40–42]. The resulting fragments are subsequently degraded by the processive 3′ to 5′ exonucleases, polynucleotide phosphorylase (PNPase), RNase II and RNase R. In vitro analyses of these exonucleases reveal that they do not efficiently process RNA species less than ~5-nt in length. Specifically, RNase R produces 2-nt and 3-nt terminal fragments in vitro 43, 44, RNase II produces 3- to 5-nt terminal fragments 45, 46, while PNPase produces 2- to ~5-nt terminal fragments 47. Thus, degradation of a single full-length transcript in vivo likely results in the production of several 2- to ~5-nt nanoRNAs as the terminal products of digestion by PNPase, RNaseII or RNase R. Furthermore, as the processing of the fragments produced initially by RNase E cleavage occurs in a 3′ to 5′ manner, nanoRNAs that result from RNA degradation are likely derived from the 5′ ends of the initial fragments derived from RNase E cleavage. Thus, the positions where RNase E cleaves a particular mRNA transcript would be predicted to specify the 5′end of a specific set of nanoRNAs.
The 3′ fragment resulting from cleavage with RNase E contains a 5′ monophosphate, and so any subsequent nanoRNA derived as a terminal degradation product of this fragment will also contain a 5′ monophosphate. Thus, full-length transcripts initiated with such nanoRNAs would themselves be expected to be good substrates for RNase E, as RNase E exhibits a strong preference for transcripts carrying a 5′ monophosphate 28. Although RNase E is the principal endonuclease in E. coli responsible for the majority of RNA turnover, other endonucleases are present in the cell and some of these generate a 5′ hydroxyl group upon cleavage [reviewed in 48]. Initiation with any subsequently derived 5′ hydroxyl carrying nanoRNA would result in the production of a transcript that would likely be resistant to subsequent attack by RNase E.
Products of abortive transcription initiation
NanoRNAs are likely generated in vivo during the process of “abortive initiation”. In particular, during transcription initiation, the RNAP-promoter initial transcribing complex can engage in tens or hundreds of cycles of synthesis and release of short RNA transcripts between 2 nt and ~15 nt in length [reviewed in 49, 50]. This process is called abortive initiation and the short transcripts that are released during abortive initiation are termed abortive RNA transcripts. The production of abortive transcripts is well established in reactions performed in vitro. Furthermore, it has recently been established that abortive transcripts are also produced in vivo 51.
Abortive transcripts can be produced in vast stoichiometric excess over full-length transcripts. Thus, a single RNAP-promoter complex could, in principle, produce large quantities of particular nanoRNA species. Short abortive transcripts (i.e. those between 2-nt and ~4-nt in length) would be suitable primers in the absence of any exonucleolytic processing, whereas longer abortive transcripts (i.e. those between ~5-nt and 15-nt) would require processing before becoming a size suitable for use as primers.
Products of RNA cleavage during transcription elongation
During transcription elongation RNAP can “backtrack”. Backtracking is a process of reverse translocation during which the RNAP catalytic center slides back relative to the 3′ end of the nascent transcript [reviewed in 52, 53]. Backtracking can occur due to (i) the presence of a weak RNA-DNA hybrid, (ii) an encounter between RNAP and a protein roadblock, or (iii) after the misincorporation of a nucleotide into the nascent transcript. For elongation to resume the backtracked complex must either reposition the 3′ end of the nascent transcript in the catalytic center by reversing the backward movement of the enzyme or generate a new RNA 3′ end in the active center via endonucleolytic cleavage. Endonucleolytic cleavage of the nascent transcript, which can be catalyzed by the RNAP active center alone or stimulated by the Gre proteins, will liberate a small 5′ monophosphate containing RNA transcript from the elongation complex. For example, in the case of misincorporation of a nucleotide, backtracking by one base pair precedes cleavage of the penultimate phosphodiester bond, liberating a dinucleotide 54. Therefore endonucleolytic cleavage of nascent transcripts associated with backtracked elongation complexes, in principle, may provide another potential source of nanoRNAs in vivo 55.
Products of cyclic dinucleotide degradation
Cyclic di-guanosine monophosphate (c-di-GMP) is a second messenger that has attracted considerable attention of late because of its ability to control both biofilm formation and motility in many different bacteria 56–58. C-di-GMP can influence many different cellular processes either directly or indirectly through binding specific receptor proteins or RNAs 58. For example, the nucleotide can influence gene expression by binding directly to several different transcription regulators 59, 60. Synthesized by proteins containing a so-called GGDEF domain, c-di-GMP is degraded by EAL domain-containing phosphodiesterases to the dinucleotide pGpG 61, which could function as a nanoRNA to prime transcription initiation. A second class of phosphodiesterases containing a so-called HD-GYP domain can degrade c-di-GMP to pGpG as well, but these enzymes can also degrade pGpG further to two molecules of GMP 62. Most bacteria contain genes encoding GGDEF and EAL domain-containing proteins but the number of genes specifying each of these family members can vary considerably between organisms. Because certain GGDEF and EAL domain-containing proteins are predicted to contain signal input domains, it is possible that the synthesis and breakdown of c-di-GMP by these particular proteins only occurs in response to specific environmental signals. However, there are known instances in which phenotypic and genotypic variants arise within a bacterial population that exhibit increased intracellular c-di-GMP concentrations 63–66; it is conceivable that in these instances the pGpG dinucleotide breakdown product of c-di-GMP is also more abundant and consequently that priming of transcription by pGpG is particularly pronounced. Although cyclic di-adenosine monophosphate (c-di-AMP) is produced by some bacteria 67, it remains to be learned whether this cyclic dinucleotide is broken down to pApA in the cell and could therefore serve as a source of nanoRNAs.
NanoRNA-degrading enzymes as key regulators of nanoRNA-mediated priming
Goldman et al. found that depletion of Orn in P. aeruginosa led to widespread use of nanoRNAs to prime transcription initiation. Thus, this study indicates that one major role of Orn, and presumably other nanoRNA-degrading enzymes, is to suppress nanoRNA-mediated priming of transcription. Furthermore, the studies suggest that nanoRNA degrading enzymes play a key role in regulating gene expression by influencing the extent to which transcription initiation occurs using NTPs only or a nanoRNA primer.
Different bacteria appear to have evolved distinct strategies to degrade nanoRNAs [for review see 42]. For example, E. coli appears to carry only a single nanoRNA-degrading enzyme 15, whereas Bacillus subtilis carries several proteins (all of which are unrelated to Orn) that can degrade nanoRNAs 17. The presence of one (or more) enzymes dedicated to the degradation of nanoRNAs, in principle, provides a facile way of altering the degradation of nanoRNAs without affecting other cellular processes.
It remains to be learned why some bacteria carry one nanoRNA-degrading enzyme whereas other bacteria carry multiple enzymes. One potential explanation may be that nanoRNA-degrading enzymes have differing substrate specificities. Thus, in the case of cells carrying multiple nanoRNA-degrading enzymes the bacteria may be able to alter the composition of the cellular nanoRNA pool by down- or up-regulating the activity of one of these enzymes while the activity of the other(s) remains unchanged. Although a systematic analysis of the substrate specificities of the currently identified nanoRNA-degrading enzymes has yet to be reported, some information regarding the substrate specificity of E. coli Orn is available. Interestingly, when tested against a series of dinucleotides, E. coli Orn was least active on a GpG dinucleotide 68, which as mentioned above, may accumulate in cells under conditions where c-di-GMP levels are elevated.
Loss of Orn function in E. coli leads to a loss of viability 15. This loss of viability can be rescued by the introduction of a heterologous nanoRNA-degrading enzyme into cells 16, 17. These findings suggest that the accumulation of nanoRNAs is toxic in E. coli. The basis for the toxic effects of nanoRNA accumulation in E. coli remains to be determined. However, one possibility, as proposed by Goldman et al., is that the toxic effects of nanoRNAs are a consequence of altered gene expression due to nanoRNA-mediated priming.
In contrast to the situation in E. coli, Orn is apparently not essential in either P. aeruginosa 18 or Streptomyces 69 (although loss of Orn in Streptomyces does lead to a growth defect). One possible explanation for the fact that P. aeruginosa and Streptomyces remain viable in the absence of Orn function is that these organisms may carry some as of yet unidentified nanoRNA-degrading enzyme that allows for survival in the absence of Orn. However, this explanation seems unlikely given the demonstration that nanoRNAs accumulate in P. aeruginosa upon depletion of Orn. An alternative, and perhaps more likely explanation, is that the accumulation of nanoRNAs is not lethal in these organisms.
Summary/future prospects
Goldman et al. demonstrate that artificial depletion of Orn allows widespread nanoRNA-mediated priming of transcription coupled with global alterations in gene expression to occur in bacterial cells. These findings raise the possibility that nanoRNAs represent a new class of regulatory small RNAs that can influence gene expression through a novel mechanism—by being incorporated directly into a target transcript as opposed to hybridizing to it. Nevertheless, the extent to which nanoRNA-mediated priming occurs in wild-type cells of P. aeruginosa, or in cells of any other bacterium, is still an unknown. Furthermore, if nanoRNA-mediated priming does occur in wild-type cells, it need not necessarily play a specific regulatory function; rather, it may influence the expression of certain genes in a stimulus-independent manner, thus helping to shape the transcriptome. However, because the production or activity of nanoRNA-degrading enzymes such as Orn could in principle be modulated, and because the potential sources of nanoRNAs could differ depending on the particular growth conditions, it is conceivable that nanoRNA-mediated priming of transcription could be more prevalent under certain growth conditions than others, or more prevalent in some bacteria, than in others.
The study by Goldman et al. demonstrates that at least one diagnostic feature of nanoRNA-mediated priming is an extension at the 5′ end of a transcript, or, put another way, a change in the transcription start point to a more upstream position. In principle, the high-throughput sequencing approaches outlined in the study by Goldman et al., which relied upon the analysis of the 5′ ends of all transcripts regardless of their phosphorylation status, could be used to investigate the extent of nanoRNA-mediated priming in wild-type cells under different growth conditions. Importantly, the study by Goldman et al. also suggests that any approach for the identification of transcription start points that relies upon the analyses of only those transcripts carrying a 5′ triphosphate [see, for example, 70, 71] might miss a significant portion of transcripts whose synthesis is primed with a nanoRNA.
In conclusion, the study by Goldman et al. provides considerable impetus for addressing the extent to which nanoRNA-mediated priming occurs in bacteria and, if so, whether nanoRNA-mediated priming plays a specific regulatory role or serves simply to maintain the expression state of a given set of genes under all conditions.
Acknowledgments
We thank Ann Hochschild for comments on the manuscript. This work was funded by grant GM096454 from the NIH (to B.E.N. and S.L.D.) and a Pew Scholars Award to B.E.N.
References
- 1.Hurwitz J. The discovery of RNA polymerase. J Biol Chem. 2005;280:42477–42485. doi: 10.1074/jbc.X500006200. [DOI] [PubMed] [Google Scholar]
- 2.Niyogi SK, Stevens A. Studies on ribonucleic acid polymerase with synthetic polyribonucleotides as templates: effect of oligonucleotides on the reactions. Biochem Biophys Res Commun. 1964;16:272–277. doi: 10.1016/0006-291x(64)90339-0. [DOI] [PubMed] [Google Scholar]
- 3.Niyogi SK, Stevens A. Studies of the Ribonucleic Acid Polymerase from Escherichia Coli.Iv.Effect of Oligonucleotides on the Ribonucleic Acid Polymerase Reaction with Synthetic Polyribonucleotides as Templates. J Biol Chem. 1965;240:2593–2598. [PubMed] [Google Scholar]
- 4.Learned RM, Tjian R. In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J Mol Appl Genet. 1982;1:575–584. [PubMed] [Google Scholar]
- 5.Wilkinson JK, Sollner-Webb B. Transcription of Xenopus ribosomal RNA genes by RNA polymerase I in vitro. J Biol Chem. 1982;257:14375–14383. [PubMed] [Google Scholar]
- 6.Samuels M, Fire A, Sharp PA. Dinucleotide priming of transcription mediated by RNA polymerase II. J Biol Chem. 1984;259:2517–2525. [PubMed] [Google Scholar]
- 7.Reich C, Zeller M, Milkereit P, Hausner W, Cramer P, Tschochner H, Thomm M. The archaeal RNA polymerase subunit P and the eukaryotic polymerase subunit Rpb12 are interchangeable in vivo and in vitro. Mol Microbiol. 2009;71:989–1002. doi: 10.1111/j.1365-2958.2008.06577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodny GM. The regulation of gene expression in eukaryotic cells. Med Hypotheses. 1975;1:15–22. doi: 10.1016/0306-9877(75)90036-5. [DOI] [PubMed] [Google Scholar]
- 9.Robertson HD, Dickson E. RNA processing and the control of gene expression. Brookhaven Symp Biol. 1975:240–266. [PubMed] [Google Scholar]
- 10.Downey KM, So AG. Studies on the kinetics of ribonucleic acid chain initiation and elongation. Biochemistry. 1970;9:2520–2525. doi: 10.1021/bi00814a019. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman DJ, Niyogi SK. RNA initiation with dinucleoside monophosphates during transcription of bacteriophage T4 DNA with RNA polymerase of Escherichia coli. Proc Natl Acad Sci U S A. 1973;70:574–578. doi: 10.1073/pnas.70.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. NanoRNAs prime trancription initiation in vivo. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.06.005. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niyogi SK, Datta AK. A novel oligoribonuclease of Escherichia coli. I. Isolation and properties. J Biol Chem. 1975;250:7307–7312. [PubMed] [Google Scholar]
- 14.Yu D, Deutscher MP. Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli. J Bacteriol. 1995;177:4137–4139. doi: 10.1128/jb.177.14.4137-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007;35:4552–4561. doi: 10.1093/nar/gkm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang M, Zeisberg WM, Condon C, Ogryzko V, Danchin A, Mechold U. Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis. Nucleic Acids Res. 2009;37:5114–5125. doi: 10.1093/nar/gkp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minkley EG, Pribnow D. Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J Mol Biol. 1973;77:255–277. doi: 10.1016/0022-2836(73)90335-5. [DOI] [PubMed] [Google Scholar]
- 20.Di Nocera PP, Avitabile A, Blasi F. In vitro transcription of the Escherichia coli histidine operon primed by dinucleotides. Effect of the first histidine biosynthetic enzyme. J Biol Chem. 1975;250:8376–8381. [PubMed] [Google Scholar]
- 21.Smagowicz WJ, Scheit KH. Primed abortive initiation of RNA synthesis by E. coli RNA polymerase on T7 DNA. Steady state kinetic studies. Nucleic Acids Res. 1978;5:1919–1932. doi: 10.1093/nar/5.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grachev M, Zaychikov E, Ivanova E, Komarova N, Kutyavin I, Sidelnikova N, Frolova I. Oligonudeotides complementary to a promoter over the region −8…+ 2 as transcription primers for E. coli RNA polymerase. Nucleic Acids Res. 1984;12:8509–8524. doi: 10.1093/nar/12.22.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruetsch N, Dennis D. RNA polymerase. Limit cognate primer for initiation and stable ternary complex formation. J Biol Chem. 1987;262:1674–1679. [PubMed] [Google Scholar]
- 24.Turnbough C, Switzer R. Regulation of Pyrimidine Biosynthetic Gene Expression in Bacteria: Repression without Repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smagowicz W, Scheit K. A minimal mechanism for abortive initiation of transcription of T7 DNA. Nucleic Acids Res. 1981;9:5845–5854. doi: 10.1093/nar/9.21.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perrin DM, Chen C-hB, Xu Y, Pearson L, Sigman DS. Gene-Specific Transcription Inhibitors. Pentanucleotides Complementary to the Template Strand of Transcription Start Sites. J Am Chem Soc. 1997;119:5746–5747. [Google Scholar]
- 27.Milne L, Xu Y, Perrin DM, Sigman DS. An approach to gene-specific transcription inhibition using oligonucleotides complementary to the template strand of the open complex. Proc Natl Acad Sci U S A. 2000;97:3136–3141. doi: 10.1073/pnas.050544597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 29.Deana A, Celesnik H, Belasco J. The bacterial enzyme RppH triggers messenger RNA degradation by 5prime pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 30.Goliger JA, Yang XJ, Guo HC, Roberts JW. Early transcribed sequences affect termination efficiency of Escherichia coli RNA polymerase. J Mol Biol. 1989;205:331–341. doi: 10.1016/0022-2836(89)90344-6. [DOI] [PubMed] [Google Scholar]
- 31.Telesnitsky AP, Chamberlin MJ. Sequences linked to prokaryotic promoters can affect the efficiency of downstream termination sites. J Mol Biol. 1989;205:315–330. doi: 10.1016/0022-2836(89)90343-4. [DOI] [PubMed] [Google Scholar]
- 32.Sorensen KI, Neuhard J. Dual transcriptional initiation sites from the pyrC promoter control expression of the gene in Salmonella typhimurium. Mol Gen Genet. 1991;225:249–256. doi: 10.1007/BF00269856. [DOI] [PubMed] [Google Scholar]
- 33.Wilson HR, Archer CD, Liu JK, Turnbough CL., Jr Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J Bacteriol. 1992;174:514–524. doi: 10.1128/jb.174.2.514-524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi F, Turnbough CL., Jr Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J Mol Biol. 1995;254:552–565. doi: 10.1006/jmbi.1995.0638. [DOI] [PubMed] [Google Scholar]
- 35.Belasco JG. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol. 2010;11:467–478. doi: 10.1038/nrm2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celesnik H, Deana A, Belasco J. Initiation of RNA Decay in Escherichia coli by 5prime Pyrophosphate Removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tock MR, Walsh AP, Carroll G, McDowall KJ. The CafA protein required for the 5prime-maturation of 16 S rRNA is a 5prime-end-dependent ribonuclease that has context-dependent broad sequence specificity. J Biol Chem. 2000;275:8726–8732. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- 38.Jiang X, Belasco JG. Catalytic activation of multimeric RNase E and RNase G by 5prime-monophosphorylated RNA. Proc Natl Acad Sci U S A. 2004;101:9211–9216. doi: 10.1073/pnas.0401382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClure WR, Cech CL, Johnston DE. A steady state assay for the RNA polymerase initiation reaction. J Biol Chem. 1978;253:8941–8948. [PubMed] [Google Scholar]
- 40.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Condon C. Maturation and degradation of RNA in bacteria. Curr Opin Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Condon C. Molecular Biology of RNA Processing and Decay in Prokaryotes. Academic Press; London, UK: 2009. [Google Scholar]
- 43.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 44.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 45.Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J Mol Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 46.Amblar M, Barbas A, Gomez-Puertas P, Arraiano CM. The role of the S1 domain in exoribonucleolytic activity: substrate specificity and multimerization. RNA. 2007;13:317–327. doi: 10.1261/rna.220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Z, Yang WZ, Lin-Chao S, Chak KF, Yuan HS. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA. 2008;14:2361–2371. doi: 10.1261/rna.1244308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennell D. Processing endoribonucleases and mRNA degradation in bacteria. J Bacteriol. 2002;184:4645–4657. doi: 10.1128/JB.184.17.4645-4657.2002. discussion 4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu LM. Promoter clearance and escape in prokaryotes. Biochimica et biophysica acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 50.Hsu LM. Monitoring abortive initiation. Methods. 2009;47:25–36. doi: 10.1016/j.ymeth.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochimica et biophysica acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 53.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 54.Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 55.Toulme F, Mosrin-Huaman C, Sparkowski J, Das A, Leng M, Rahmouni AR. GreA and GreB proteins revive backtracked RNA polymerase in vivo by promoting transcript trimming. EMBO J. 2000;19:6853–6859. doi: 10.1093/emboj/19.24.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 58.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 59.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryan RP, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Beyhan S, Yildiz FH. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol Microbiol. 2007;63:995–1007. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- 64.Meissner A, Wild V, Simm R, Rohde M, Erck C, Bredenbruch F, Morr M, Romling U, Haussler S. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ Microbiol. 2007;9:2475–2485. doi: 10.1111/j.1462-2920.2007.01366.x. [DOI] [PubMed] [Google Scholar]
- 65.Starkey M, et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C, Kaever V, Landmann R, Jenal U. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 2010;6:e1000804. doi: 10.1371/journal.ppat.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Romling U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci Signal. 2008;1:pe39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- 68.Datta AK, Niyogi K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J Biol Chem. 1975;250:7313–7319. [PubMed] [Google Scholar]
- 69.Ohnishi Y, Nishiyama Y, Sato R, Kameyama S, Horinouchi S. An oligoribonuclease gene in Streptomyces griseus. J Bacteriol. 2000;182:4647–4653. doi: 10.1128/jb.182.16.4647-4653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]