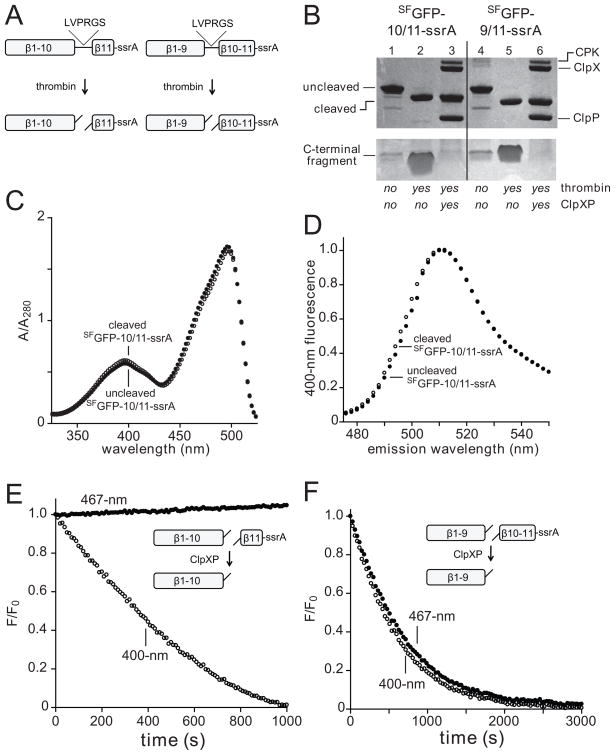
Fig. 2.
(A) A thrombin cleavage sequence (LVPRGS) was inserted between strands 10/11 or 9/10 in the SFGFP-10/11-ssrA and SFGFP-9/10-ssrA proteins (NCBI accession codes JF951868 and JF951869, respectively), allowing creation of split proteins. (B) SDS-PAGE showing SFGFP-10/11-ssrA or SFGFP-9/10-ssrA (10 μM each) before and after thrombin cleavage and ClpXP (0.3 μM ClpX6; 0.9 μM ClpP14) extraction/degradation. The gel is a composite, with the lower portion taken from a gel containing 8-fold more sample than the upper portion. CPK, creatine phosphokinase. (C) Absorbance spectra of SFGFP-10/11-ssrA before (closed circles) or after (open circles) thrombin cleavage. (D) Fluorescence emission spectrum of SFGFP-10/11-ssrA before (closed circles) or after (open circles) thrombin cleavage. (E) Incubation of 10 μM thrombin-cleaved SFGFP-10/11-ssrA with 1 μM ClpXP (1 μM ClpX6; 2 μM ClpP14) and 4 mM ATP resulted in loss of 400-nm (open circles) but not 467-nm (closed circles) fluorescence. (F) Incubation of 10 μM thrombin-cleaved SFGFP-9/10-ssrA with 1 μM ClpXP and 4 mM ATP resulted in loss of 400-nm (open circles) and 467-nm (closed circles) fluorescence. The experiments in panels E and F contained an ATP-regeneration system.