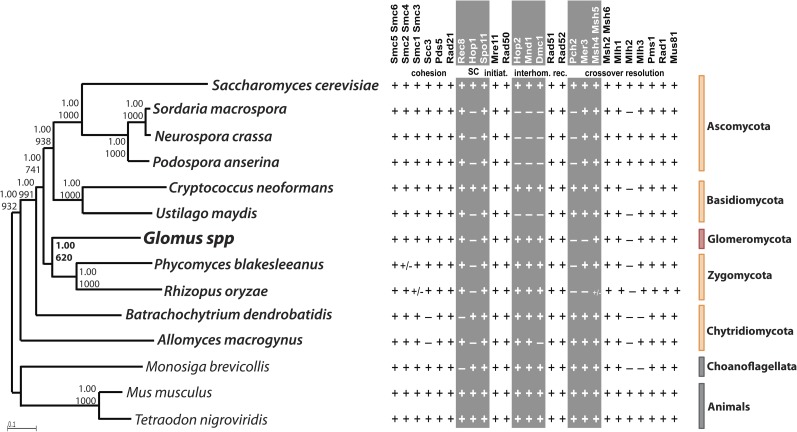
FIG. 2.
Phylogenetic tree of 12 concatenated DNA repair protein sequences (Exo1, Rad1, Mlh1, Mre11, Msh6, Mus81, Smc6, Top1, Top3, Rad23, Rad50, Rad52) (left) and list of the core meiotic genes (right). Left side of the figure: Glomus spp. (G. irregulare and G. diaphanum) are highlighted in red, all surveyed fungi (Ascomycota, Basidiomycota, Chytridiomycota, and Zygomycota) in orange, and outgroup organisms (one choanoflagellate and two animals) in gray (supplementary table S2, Supplementary Material online). Numbers at nodes correspond to Bayesian posterior probabilities (top) and bootstrap supports from 1,000 replicates of maximum likelihood (PhyML) analysis (bottom). Scale bar represents 0.1 amino acid substitutions per site. Right side of the figure: list of the core meiotic proteins (adapted from references San-Segundo and Roeder 1999; Villeneuve and Hillers 2001; Malik et al. 2008; Joshi et al. 2009 and references therein) and their presence (+) and absence (−, i.e., not detected) in the fungal genomes surveyed in this study. +/− denotes the absence of the given genes in some species belonging to that specific phylum. Meiosis-specific proteins are shown in gray columns. A. Ascomycota; B. Basidiomycota; C. Chytridiomycota; Z. Zygomycota; G. Glomeromycota (i.e., AMF). Orthologs of Rad21, Rad51, Pms1, and Mlh and meiosis-specific Spo11-1, Rec8, Hop1, Hop2, Mnd1, and Dmc1 genes of basidiomycetes and B. dendrobatidis were identified with assistance from Arthur Pightling.