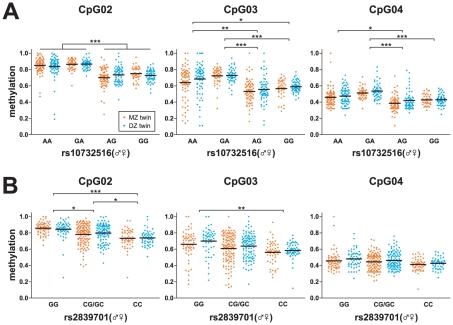
Figure 6. Influence of rs10732516 and rs2839701 on the DNA methylation levels in the H19-ICR region.
(A) DNA methylation results are sorted on the rs10732516 genotype, where the parent of origin is also taken into account. The rs10732516 [AA] and [GA] genotypes ([♂♀]) display elevated DNA methylation levels compared to [AG] and [GG] genotypes. These differences in DNA methylation are visible throughout the H19-ICR locus but are most apparent for CpG sites 2, 3 and 4. (B) Similarly, DNA methylation results are sorted based on the rs2839701 genotype. For CpG sites 2, 3 and 4 in the H19-ICR region, the rs2839701 [G] allele is associated with a higher DNA methylation compared to the [C] allele. These results show that both polymorphisms within the IGF2/H19 region may predict or influence the DNA methylation status in individuals. Significant differences between groups were identified using the non-parametric Kruskal-Wallis test, followed by a Dunn's multiple comparison test (*: P-value<0.05; **: P-value<0.01; ***: P-value<0.001).