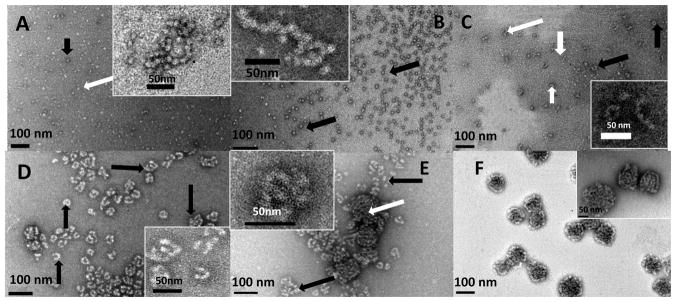
Figure 2.
Representative TEM images of amelogenin microstructures resulting from co-assembly of rP172 and its proteolytic products during proteolysis in Tris-Cl buffer, pH 8.0, at 37 °C, collected at 0.1 hr (A), 1 hr (B), 2 hrs (C), 4 hrs (D), 10 hrs (E) and 20 hrs (F). (A) The lightly stained rP172 particles (white arrow) are connected with some deeply stained spherical particles (black arrow), forming a beaded-like structure, the inset presenting magnification of typical chains; (B) At 1 hour proteolysis, some tightly associated and elongated chain-like microstructures (inset) are formed within the uniform suspension of spherical particles (black arrows); (C) After 2 hours proteolysis, cylindrical-shaped nanorods (white arrows) formed and some of nanorods curved into O-like or U-shaped microstructures (black arrows). Inset shows typical images of nanorods and their curled microstructures at higher magnification; (D) After 4 hours proteolysis, different morphologies composed of nanorods were formed (black arrows); (E) After 10 hours proteolysis, a new mixture of compact aggregates (white arrows) were detected among the remaining rod-like structures (black arrows). Inset presents a typical morphology at higher magnification; (F) After 20 hours of proteolysis, the tightly compact ball-like aggregates grew larger, concomitant with disappearance of rod-like morphologies. Inset presents a typical morphology at higher magnification. The analyses of all the particles sizes are described in the Table 1.