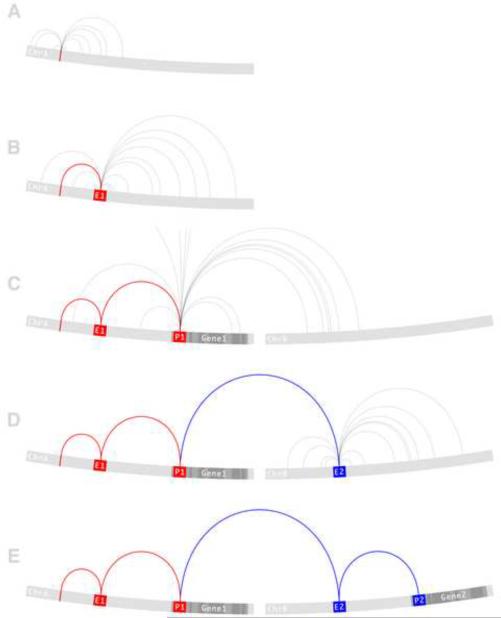
Figure 1.
A proposed computational framework to predict functional consequences of genetic variation. The red edges track the path from a disease-associated variant to a disease-associated gene. The blue edges track the path from the disease-associated gene to a functional target. (a) A variant node on chromosome A (red) has been associated with a disease. The variant may be causal or may be serving as a proxy of nearby causal variant so the first set of edges connect to those variant nodes that are in linkage disequilibrium (LD) with the associated variant. (b) One of the LD edges connects to the E1 enhancer node. Because an enhancer is a functional entity that directly regulates gene transcription, the second set of edges connect the E1 enhancer node to its long-range interacting partners. (c) One of the long-range interactions connects the E1 enhancer node to the P1 promoter node of the Gene1 gene node. This path reflects known empirical evidence in support of a regulatory interaction between the E1 enhancer and Gene1 mediated by the P1 promoter. Gene1 is a known transcription factor but has not previously been linked to the disease. (d) The third set of edges connect the transcription factor Gene1 to all of its known binding sites. One of these transcription factor binding site edges connects Gene1 to the E2 enhancer node found on chromosome B. Continuing the pattern established here, a fourth set of edges are drawn to connect the E2 enhancer node to its long-range interacting partners. (e) One of these long-range interactions connects the E2 enhancer node to the P2 promoter node of the Gene2 gene node. Gene2 is known to affect the disease. This final link creates a complete path through the GWRN from the associated variant to the causal gene. Every edge along this path is supported by empirical evidence however, this process is meant to provide the most likely causal explanation given the available data and as such is best used to guide the generation of novel hypotheses that can be experimentally validated.