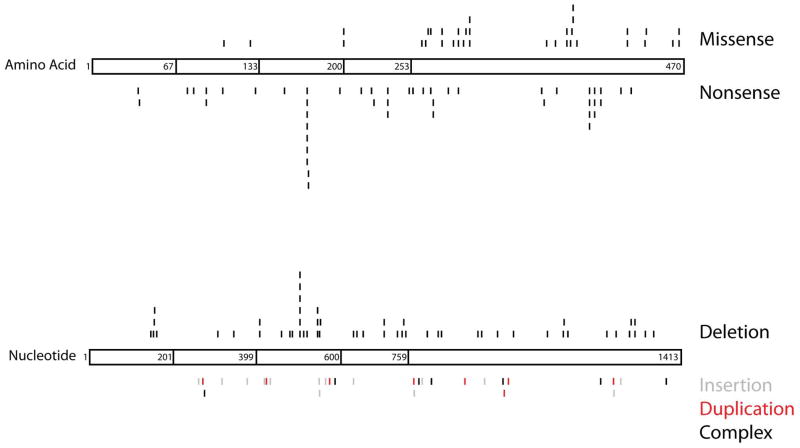
Figure 1.
Schematic of known DAX1 mutations. The DAX1 protein structure is divided into three and a half amino-terminal repeats (amino acids 1–67, 68–133, 134–200 and 201–253) and one nuclear receptor-like domain (amino acids 254–470). In the top diagram, missense (top) and nonsense (bottom) mutations are shown at specific amino acids along the length of DAX1. The bottom figure depicts deletions (black above protein), insertions (gray below protein), duplications (red below protein) and complex deletions/insertions (black below protein), which are assigned based on nucleotide number. Visualization of all naturally occurring mutations demonstrates clustering at specific points along the DAX1 protein. Numbering of mutations began at the A of the ATG translational initiation codon (Ensembl transcript ID ENST00000378970 and protein ID ENSP00000368253). The locations of the frameshift mutations (deletions, insertions, duplications and complex deletions/insertions) are designated by the first nucleotide altered. References are found above in the section “Genetics of AHC.” This represents a comprehensive list of DAX1 mutations to the best of our knowledge. Please refer to the original articles for details.