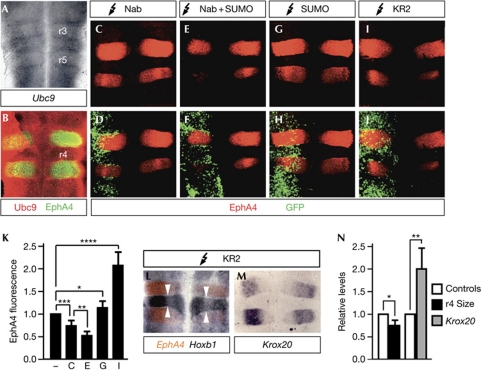
Figure 4.
Nab sumoylation regulates Krox20 target genes in vivo. Flat-mounted chick hindbrains were analysed by (A) in situ hybridization using a Ubc9 antisense RNA probe or by (B) immunofluorescence using Ubc9 and EphA4 antibodies. (C–J) The neural tube of chick embryos was electroporated with constructs expressing the proteins indicated at the top of each panel. Green fluorescent protein (GFP) was used to monitor electroporation. Electroporated hindbrains were processed for (C–J) immunofluorescence using EphA4 antibodies, or (L,M) in situ hybridization using EphA4, Hoxb1 and Krox20 RNA probes. Arrowheads in L delimit rhombomere (r) 4. Electroporations were performed on the left side of embryos. (K) Fluorescence signals of EphA4 hybridization in C, E, G and I were measured using the MetaMorph software. For that, regions of the same area encompassing r3–r5 were defined on both electroporated and control sides. (N) Size of r4 was determined by measuring the Hoxb1-positive area using the ImageJ application, and intensity of the Krox20 hybridization signal was measured with the MetaMorph software on inverted grey-scale-converted images. Relative levels were normalized with respect to those on the control side (−). Values correspond to the mean±s.d. of 5–8 samples from three independent experiments. Statistical significance was analysed using Student's t-test: (K) *P=0.13, **P<0.025, ***P<0.005 and ****P<0.001; (N) *P<0.025 and **P<0.005. SUMO, small ubiquitin-like modifier.