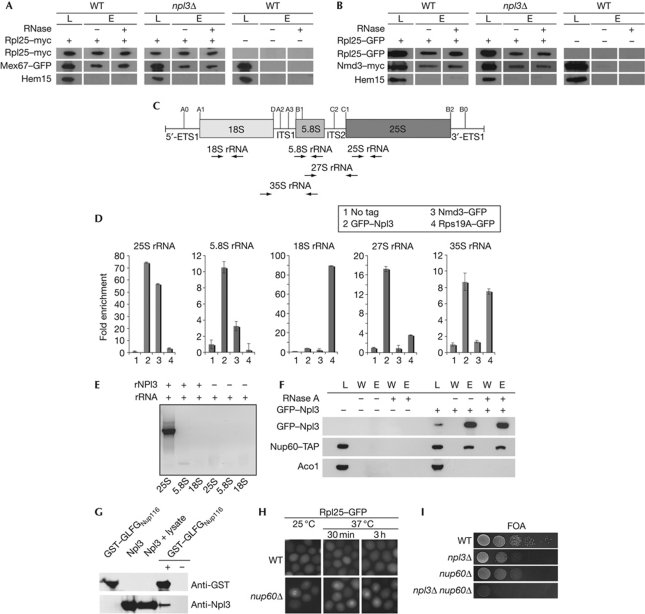
Figure 4.
Npl3 functions as an independent export adaptor for the pre-60S ribosomal subunit. (A) Deletion of NPL3 does not impair the binding of Mex67 to the large ribosomal subunit. Co-immunoprecipitations of Mex67–GFP (green fluorescent protein) with Rpl25–myc were performed in wild-type (WT) and npl3Δ cells. (B) Deletion of NPL3 does not influence the binding of Nmd3 to the pre-60S subunit as shown in co-immunoprecipitations of Nmd3–myc with Rpl25–GFP. Total lysates shown in A and B were split and incubated with or without RNase before elution. Western blot analyses with antibodies against GFP, myc and the control protein Hem15 are shown. (C) Scheme of the 35S ribosomal RNA (rRNA) precursor and location of the processing sites. The pre-rRNA encodes the 18S, 5.8S and the 25S rRNAs, which are flanked by the 5′ and 3′ external transcribed spacers (5′-ETS and 3′-ETS) and separated by internal transcribed spacers 1 and 2 (ITS1 and ITS2). From the 35S precursor, rRNA cleavage generates the 27S and the 20S pre-rRNA, from which the 5.8S+25S and the 18S rRNAs are generated, respectively. The positions for the primers used in the quantitative reverse transcription PCR (RT–PCR) are shown as arrows. (D) Npl3 binds to the 35S, 27S, 25S and 5.8S rRNA in vivo. Wild-type cells were used as ‘no tag’ control. npl3Δ carrying a CEN GFP–NPL3 plasmid, nmd3-2 carrying a CEN NMD3–GFP plasmid and genomically encoded RPS19A–GFP were grown to log phase before GFP immunoprecipitations were performed. Furthermore, the associated RNA was extracted and used for quantitative RT–PCR experiments. (E) In vitro experiments showing that Npl3 binds directly to the 25S rRNA. Regions of the 25S, 5.8S and the 18S rRNAs were in vitro transcribed and incubated with recombinant His–Npl3 (rNpl3). Co-immunoprecipitations were performed and the purified rRNA was used for RT–PCR with specific primers, shown on an agarose gel. (F) Co-immunoprecipitations of Nup60–TAP with GFP–Npl3 were performed in WT cells. Total lysates were split and incubated with or without RNase before elution. Western blot analyses with antibodies against GFP, TAP and the control protein Aco1 are shown. (G) Npl3 interacts with FG repeats in vitro. Purified recombinant His–Npl3 (rNpl3) and a glutathione S-transferase (GST)-tagged GLFG-repeat-containing region of Nup116 were investigated for an in vitro interaction. Co-purification of rNpl3 was performed with GST pull-down in the presence of whole bacterial lysate. Western blot analyses are shown with antibodies against Npl3 and GST. (H) Deletion of NUP60 leads to export defects of the large ribosomal subunit. Wild-type and nup60Δ cells were grown to log phase and shifted to 37 °C for the indicated times. Localization of Rpl25–GFP is shown. (I) npl3Δ and nup60Δ are synthetically lethal. Wild-type cells or cells deleted for either indicated gene alone or in combination are shown on 5′-fluoroorotic acid (FOA) plates that select for the loss of a covering NPL3 plasmid on growth for 3 days at 25 °C. E, eluate; L, lysate; W, wash.