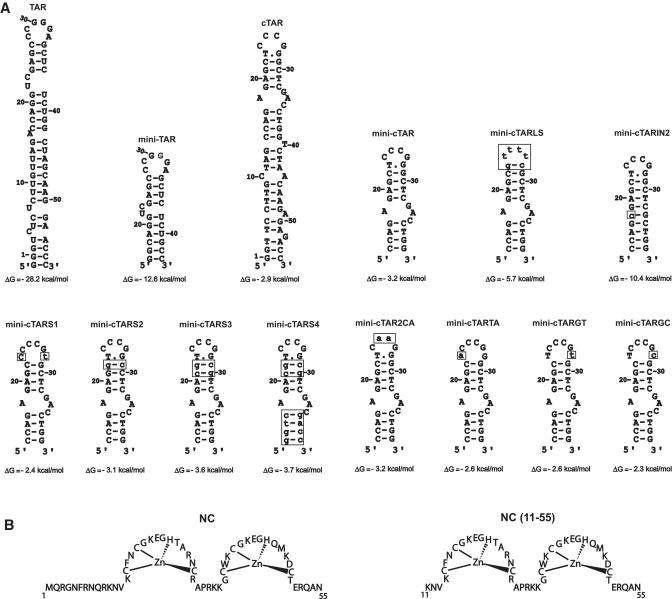
Figure 1.
(A) Predicted secondary structures for the TAR and cTAR sequences. Zuker’s folding programs (48) were used to predict the most stable secondary structures for RNAs and DNAs. The TAR RNA and cTAR DNA sequences are derived from the HIV-1 MAL isolate. Numbering is relative to the TAR and cTAR sequences. Mutations are shown as lower case letters in boxes. (B) Sequences of proteins used in this study.