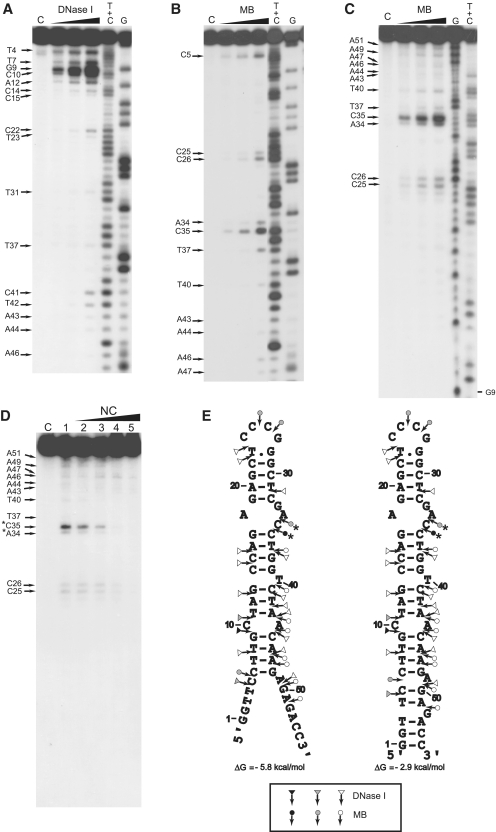
Figure 5.
Enzymatic probing of cTAR. Enzymatic probing experiments were performed as described in ‘Materials and Methods’ section. (A and B) In the absence of NC, the 3′-end-labeled cTAR DNA was incubated with DNase I (0.05, 0.1 and 0.2 U) or mung bean nuclease (MB) (1.5, 3 and 6 U). (C) In the absence of NC, the 5′-end-labeled cTAR DNA was incubated with MB (1.5, 3 and 6 U). Lanes C are controls without enzyme. G and T+C refer to Maxam–Gilbert sequence markers. Arrows indicate the cleavage sites. (D) 5′-end-labeled cTAR DNA was incubated with MB (3 U) in the absence (lane 1) or in the presence of NC (lanes 2–5). The protein to nucleotide molar ratios were 1:8 (lane 2), 1:4 (lane 3), 1:2 (lane 4) and 1:1 (lane 5). The strong protections induced by NC at the level of MB cleavage sites are indicated by asterisks. (E) Secondary structures for the partially melted and closed forms of cTAR. Delta G-values were predicted by mfold. Closed, gray and open symbols indicate strong, medium and weak cleavage sites, respectively, for the various enzymes (triangle for DNase I and circle for mung bean nuclease).