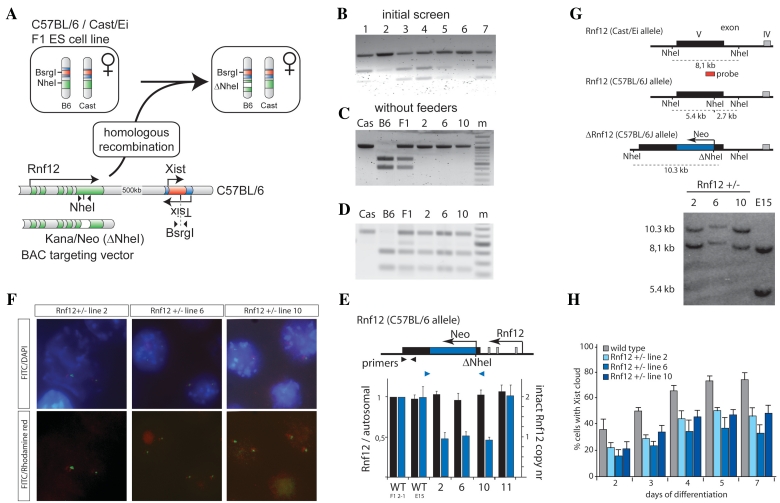
Figure 2.
Generation of Rnf12 knockout ES cell lines by disruption of the Rnf12 open reading frame. (A) Targeting of Rnf12 by insertion of a kanamycin/neomycin resistance cassette into the exon containing a NheI RFLP destroys the RFLP (present in the C57BL/6 allele). (B) PCR analysis with primers indicated in Figure 2A amplifying the NheI RFLP located in Rnf12 using genomic DNA of targeted clones 1–7. PCR fragments were digested with NheI. (C) Same as in Figure 2B, but with gDNA isolated from clones 2, 6 and 10, grown without feeder cells. Cast/Ei (Cas), C57BL/6 (B6) and Cast/Ei / C57BL/6 (F1) DNA was used as a control (m = marker). (D) Same as in Figure 2C, but PCR analysis amplifying the BsrGI RFLP located in Xist using gDNA of the targeted clones. PCR fragments were digested with BsrGI. (E) Q-PCR analysis with primers indicated using gDNA of different targeted clones. Values were normalized to values obtained with a primer set amplifying the autosomal Zfp42 gene. (F) DNA–FISH analysis with a BAC probe covering the targeting cassette (FITC), and a mix of BAC probes detecting Xist and flanking regions (Rhodamine red) (DNA stained with DAPI). (G) Southern blotting analysis, using a probe that distinguishes between the Cast/Ei (8.1 kb) and C57BL/6 (5.4 kb) alleles. A neo insertion event destroying the NheI RFLP present on the C57BL/6 allele results in a 10.3-kb fragment (clones 2, 6 and 10). (H) Three different Rnf12+/− ES cell lines and a wild-type female ES cell line were differentiated, and subjected to Xist RNA FISH. The relative number of cells showing an Xist cloud, indicative for initiation of XCI, is shown at different time points of differentiation (N > 100 per time point, error bars represent 95% confidence intervals).