Abstract
To understand the mechanism by which living cells sense mechanical forces, and how they respond and adapt to their environment, a new technology able to investigate cells behavior at sub-cellular level with high spatial and temporal resolution was developed. Thus, an atomic force microscope (AFM) was integrated with total internal reflection fluorescence (TIRF) microscopy and fast-spinning disk (FSD) confocal microscopy. The integrated system is broadly applicable across a wide range of molecular dynamic studies in any adherent live cells, allowing direct optical imaging of cell responses to mechanical stimulation in real-time. Significant rearrangement of the actin filaments and focal adhesions was shown due to local mechanical stimulation at the apical cell surface that induced changes into the cellular structure throughout the cell body. These innovative techniques will provide new information for understanding live cell restructuring and dynamics in response to mechanical force. A detailed protocol and a representative data set that show live cell response to mechanical stimulation are presented.
Protocol
1. Overview of the Integrated Microscope System
The microscope system that is used for these studies has been described in detail 1. Briefly, an inverted Olympus IX-81 microscope with TIRF attachment (Olympus, Center Valley, PA) is combined with CSU-22 Yokogawa scanning head (Yokogawa Electric Inc, Japan). A Bioscope SZ atomic force microscope (Veeco Instruments Inc, Santa Barbara, CA) is mounted on top of the optical inverted microscope, and the entire system is placed on top of a research grade optical table (Newport, irvine, CA).
The confocal assembly composed from the spinning-disk scanner, external filter wheel and its EMCCD camera (Roper Scientific-Photometrics, Tucson, AZ) are rigidly connected with an aluminum base plate placed on a silicon pad necessary for vibration damping. The confocal scanner connects with the microscope through a flange with a treaded collar that can be decoupled from the microscope during AFM measurements, such that no vibration from the spinning-disk scanner will affect the AFM measurements. An Argon-Krypton laser (Spectra Physics, Mountain View, CA) is used as excitation source for both TIRF and confocal imaging modes, and can be coupled alternatively in two optical fibers which deliver the laser to the TIRF attachment or the scanning head, respectively. Two computers control the system: Slidebook software controls the optical imaging and Nanoscope software controls the AFM.
2. Cell Culture
Live cells expressing GFP constructs targeted against specific proteins should be placed in a glass bottom cell culture dish 24 hours prior to the mechanical stimulation experiment. On the day of the experiment replace the cell culture medium with phenol red-free medium. The cell culture dish is mounted on the AFM stage with a magnetic collar.
3. AFM Tip Preparation and Microscope Set-up
The AFM tip customized with a 2 micron biotinilated glass bead will be washed five times with DPBS and then incubated 5 min with avidin (1 mg/ml), washed again with DPBS five times, and then incubated 5 min with 1 mg/mL biotinilated fibronectin. The tip will be washed five more times at the end 2, mounted on the AFM scanner, and then the protective silicon skirt will be placed around the holder. Set the microscope for video viewing to align the AFM tip in the center of the field of view. Align the optical lever by placing the laser beam at the very end of the cantilever, and then change the objective to a high magnification objective proper for single cell imaging. The spring constant of the tip has to be determined before starting the experiment. This parameter will be measured using the thermal tune method available in the Nanoscope software. Switch from video camera to EMCCD camera to allow for florescence imaging. Then, choose a cell and bring the AFM tip in contact with that specific cell. After 20 min waiting time, which is necessary for a strong focal adhesion to form at the AFM point of contact with the cell surface, the mechanical stimulation procedure will start.
4. Mechanical Stimulation of the Live Cell
TIRF imaging 3-4 will be the method of choice for visualizing and quantifying focal adhesions, and spinning-disk confocal imaging 5 will be used for visualizing the actin cytoskeleton throughout the cell body. The AFM 6 will be used in contact mode imaging under fluid.
A TIRF or confocal image of the cell will be acquired before starting the mechanical stimulation procedure. The mechanical stimulation will be induced by the functionalized AFM probe that will be moved upward in discrete steps every 3-5 min over a period of time (1-1.5 hours). The cell reactive-response will be recorded continuously in Nanoscope software as a series of height images, each line in the image representing a unit time of 1 sec. While acquiring the AFM data, the operator monitors at all times the z-position of the AFM tip to determine the proper timing for the next pull.
After each mechanical stimulation it is possible to acquire a TIRF or confocal image of the cell in Slidebook software, while continuously acquiring AFM data. The optical images can be processed off-line to create movies that show actin cytoskeleton and focal adhesions reorganization. Also, each image can be processed quantitatively to determine cell area, specific protein area, and analyze the general cell morphology.
At the end of the experiment the AFM tip is withdrawn from the cell and AFM tip sensitivity will be measured in the Nanoscope software.
The experiment is very noise sensitive! Any talking or microscope touching during the experiment should be avoided. To minimize the noise in the AFM measurements, the spinning-disk confocal should be de-coupled from the microscope. The waiting time to form a strong focal adhesion between cell surface and the AFM tip may vary depending on the cell type used and the matrix coated on the tip.
5. Representative Results
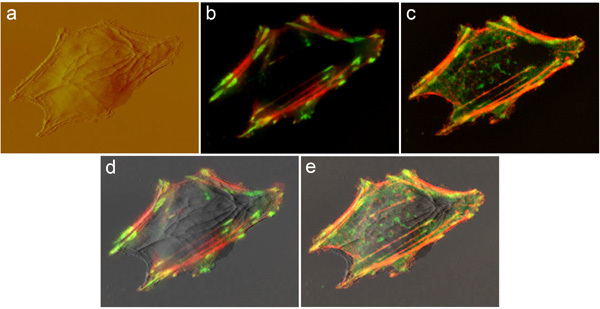 Figure 1. Same vascular smooth muscle cell (VSMC) transfected with actin-mRFP and vinculin-GFP was imaged by: (A) AFM at the apical cell surface; (B) TIRF at the basal cell surface; and (C) spinning-disk confocal microscopy throughout the cell body. The confocal image is shown as maximum projection of a 3-D stack. Good overlaps between the AFM and TIRF or spinning-disk confocal images are shown in (D) and (E), respectively. Image size is 64 x 50 micron.
Figure 1. Same vascular smooth muscle cell (VSMC) transfected with actin-mRFP and vinculin-GFP was imaged by: (A) AFM at the apical cell surface; (B) TIRF at the basal cell surface; and (C) spinning-disk confocal microscopy throughout the cell body. The confocal image is shown as maximum projection of a 3-D stack. Good overlaps between the AFM and TIRF or spinning-disk confocal images are shown in (D) and (E), respectively. Image size is 64 x 50 micron.
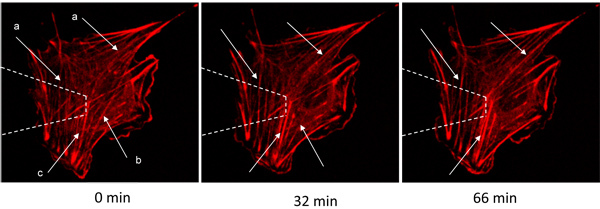 Figure 2. A VSMC transfected with actin-mRFP was mechanically stimulated by the AFM and imaged by spinning-disk confocal microscopy. The arrows indicate actin fibers that: (a) polymerize/thicken, (b) depolymerize/disappear, or (c) bundle together due to actin restructuring. White dashed lines represent the AFM tip above the cell. Image size 91 x 91 micron.
Figure 2. A VSMC transfected with actin-mRFP was mechanically stimulated by the AFM and imaged by spinning-disk confocal microscopy. The arrows indicate actin fibers that: (a) polymerize/thicken, (b) depolymerize/disappear, or (c) bundle together due to actin restructuring. White dashed lines represent the AFM tip above the cell. Image size 91 x 91 micron.
Discussion
Cytoskeleton 7-10 and focal adhesions 11-16 are dynamic structures that assemble, disperse and turn over as cells migrate or respond to mechanical force 17. They have significant functions in regulating cell growth, survival, and gene expression. Knowing their specific distribution and dynamics provides a better understanding of how cells communicate with the matrix and between themselves.
This new microscope system is broadly applicable across a wide range of molecular dynamic studies in any adherent live cells. The integrated microscope enables quantitative monitoring of rapid sub-cellular changes and provides an important novel tool for understanding the fundamental process of cellular remodeling in response to mechanical force.
Disclosures
No conflicts of interest declared.
Acknowledgments
Vinculin-GFP plasmid was a gift from Kenneth Yamada, National Institutes of Health, National Institute of Dental and Craniofacial Research, Bethesda, Maryland, and actin-mRFP plasmid was a gift from Michael Davidson, Florida State University, Tallahassee, Florida.
This work was supported by NSF Career award #0747334 and AHA-National SDG #0835205N to Andreea Trache.
References
- Trache A, Lim SM. Integrated microscopy for real-time imaging of mechanotransduction studies in live cells. J Biomed Opt. 2009;14:034024–03. doi: 10.1117/1.3155517. [DOI] [PubMed] [Google Scholar]
- Trache A, Meininger GA. Atomic Force Microscopy. In: Coico R, Kowalik T, Quarles JM, Stevenson B, Taylor RK, editors. Current Protocols in Microbiology. Wiley & Sons Inc; 2008. pp. 2.1–2.17. Volume 2C. [Google Scholar]
- Axelrod D. Selective imaging of surface fluorescence with very high aperture microscope objectives. J Biomed Opt. 2001;6:6–13. doi: 10.1117/1.1335689. [DOI] [PubMed] [Google Scholar]
- Trache A, Meininger GA. Total Internal Reflection Fluorescence Microscopy. In: Coico R, Kowalik T, Quarles JM, Stevenson B, Taylor RK, editors. Current Protocols in Microbiology. Wiley & Sons Inc; 2008. pp. 2.1–2.22. 2B. [DOI] [PubMed] [Google Scholar]
- Inoue S, Inoue T. Direct-view high-speed confocal scanner: the CSU-10. Methods Cell Biol. 2002;70:87–127. doi: 10.1016/s0091-679x(02)70003-4. [DOI] [PubMed] [Google Scholar]
- Binnig G, Quate CF, Gerber CH. Atomic force microscope. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Stamenović D. Cytoskeletal mechanics in airway smooth muscle cells. Respir Physiol Neurobiol. 2008;163:25–32. doi: 10.1016/j.resp.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Gunst SJ, Tang DD, Opazo Saez A. Cytoskeletal remodeling of the airway smooth muscle cell: a mechanism for adaptation to mechanical forces in the lung. Respir Physiol Neurobiol. 2003;137:151–168. doi: 10.1016/s1569-9048(03)00144-7. [DOI] [PubMed] [Google Scholar]
- Worth NF, Rolfe BE, Song J, Campbell GR. Vascular smooth muscle cell phenotypic modulation in culture is associated with reorganization of contractile and cytoskeletal proteins. Cell Motility and the Cytoskeleton. 2001;49:130–145. doi: 10.1002/cm.1027. [DOI] [PubMed] [Google Scholar]
- Romer HR, Birukov KG, Garcia JGN. Focal Adhesions: paradigm for a signaling nexus. Circulation Research. 2006;98:606–616. doi: 10.1161/01.RES.0000207408.31270.db. [DOI] [PubMed] [Google Scholar]
- Opazo Saez A, Zhang W, Wu Y, Turner CE, Tang DD, Gunst SJ. Tension development during contractile stimulation of smooth muscle requires recruitment of paxillin and vinculin to the membrane. Am J Physiol Cell Physiol. 2004;286:C433–C447. doi: 10.1152/ajpcell.00030.2003. [DOI] [PubMed] [Google Scholar]
- Alenghat FJ, Ingber DE. Mechanotransduction: all signals point to cytoskeleton, matrix and integrins. Sciences STKE. 2002;119:pe6–pe6. doi: 10.1126/stke.2002.119.pe6. [DOI] [PubMed] [Google Scholar]
- Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- Geiger B, Bershdsky A. Assembly and mechanosensory function of focal contacts. Curr Opin Cell Biol. 2001;13:584–592. doi: 10.1016/s0955-0674(00)00255-6. [DOI] [PubMed] [Google Scholar]
- Sastry SK, Burridge K. Focal Adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp Cell Res. 2000;261:25–36. doi: 10.1006/excr.2000.5043. [DOI] [PubMed] [Google Scholar]
- Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev Cell. 2006;10:11–20. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]


