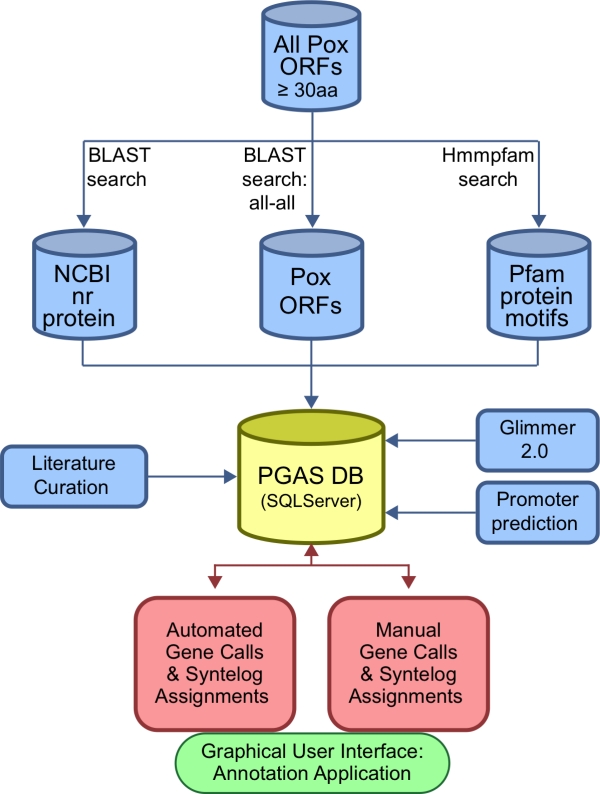
Figure 2.
The Poxvirus Genome Annotation System (PGAS) design. The PGAS pipeline (blue) automatically runs the underlying analyses in parallel on a local high-performance computing cluster for each new genome. Results from those analyses are then loaded into the PGAS database (yellow). The process of making gene calls (red) is directed from a desktop java GUI application (green).