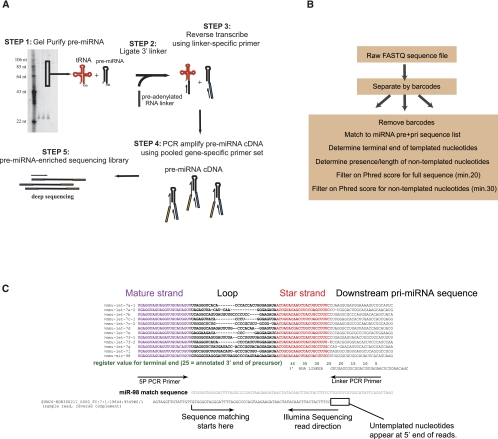
FIGURE 1.
Precursor-seq as a method of analyzing miRNA precursors. (A) Overview of experimental protocol for precursor-seq. See text for details. (B) Overview of bioinformatic analysis for precursor-seq. See text for details. (C) Sequence match algorithm overview. An alignment of the Let-7 family is shown, with mature strand in violet and star strand in red. The 3′ RNA linker is shown and both PCR primer sites. The match sequence for miR-98 is shown aligned with a sample read. Sequence matching starts at the indicated arrow and proceeds to the 3P arm. The terminal end of the precursor is determined by the last nucleotide adjacent to the RNA linker sequence. The terminal end is defined by “register,” shown in green, which corresponds to nucleotide position based on the genomic sequence. A value of “25” is defined based on the predicted terminal end in miRbase, if annotated. If not annotated, the end is defined on the expected overhang from the Drosha reaction. All registers were adjusted if necessary for the most abundant nonmodified terminal end.