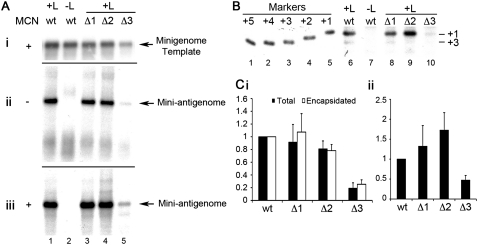
FIGURE 2.
Impact of deleting the 3′ terminal 1–3 nt of the minigenome template on RSV RNA replication in HEp-2 cells. (A) Northern blot analysis of minigenome RNAs: nuclease-resistant negative-sense minigenome templates expressed by MVA-T7 polymerase (i); positive-sense replication products expressed by the RSV polymerase from the minigenome templates in total RNA samples (ii); and positive-sense, nuclease-resistant replication products (iii). (Lane 2) A negative control in which the L plasmid, which encodes the enzymatic activity of the RSV polymerase complex, was omitted from the wild-type (wt) minigenome transfection. (B) Primer extension analysis of RNA generated from wt, Δ1, Δ2, and Δ3 templates (lanes 6,8–10). (Lane 7) A negative control in which L was omitted from the wt minigenome transfection. The band that can be detected at the top of the gel in these lanes (and in subsequent figures) is a nonspecific background band. Molecular weight markers in lanes 1–5 are end-labeled oligonucleotides, representing products initiated from position +5 to +1 of the wt template, respectively. (C) Quantitation of total (black bars) and nuclease-resistant RNA (white bars) expressed from each minigenome, as determined by Northern blotting (i); and quantitation of RNA initiated at the +1 position (relative to the wt minigenome) from each minigenome, as determined by primer extension (ii). Each value was calculated relative to the wt value, and each error bar represents standard error of the mean of at least three independent experiments.