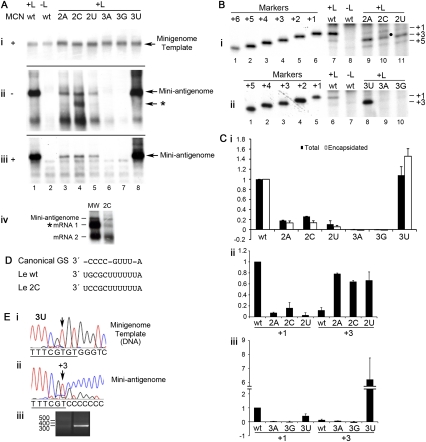
FIGURE 5.
Impact of substituting nucleotides 2G and 3C of the minigenome template on RSV RNA replication. Minigenome replication was reconstituted in HEp-2 cells simultaneously infected with MVA-T7, and the resulting RNA was analyzed. (A–C) The panels are arranged similarly to Figure 2 except that in B, the primer extensions of the RNA produced from the position 2 and position 3 substitution mutants are separated into two panels, i and ii, respectively, and (C) quantitation of the RNA initiated at +1 and +3 from the position 2 and position 3 substitution mutant templates are separated into two panels, ii and iii, respectively. (Aiv) Northern blot analysis of positive-sense RNA generated from a transcription-competent minigenome, to act as a molecular weight marker (MW), and the 2C mutant template, as indicated. The positions of the antigenome, CAT 1, and CAT 2 mRNAs are indicated, and the subgenomic RNA generated from the 2C minigenome is indicated by an asterisk in panels Aii and Aiv. In Bi the position of the +2 band detected from the 2C minigenome is indicated with a dot. (D) Sequence alignment showing the similarity between Le nucleotides 3–12 and the gene start (GS) signal sequence. (E) 5′-RACE sequence analysis of the RNA products generated from the 3U mutant template; arranged as Figure 3C.