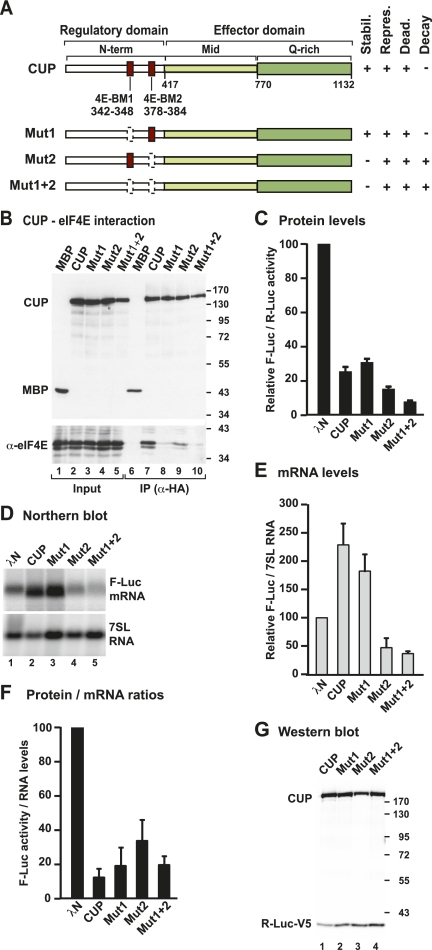
Figure 2.
CUP represses mRNA expression independently of eIF4E binding. (A) Domain organization of CUP protein. CUP contains an N-terminal regulatory domain (N-term) containing two eIF4E-binding motifs (4E-BM1 and 4E-BM2), a middle region (Mid), and a glutamine-rich C-terminal region (Q-rich). The Mid and Q-rich regions define the effector domain. The bottom panel shows the CUP point mutants used in this study and a summary of their activities. (stabil.) mRNA stabilization; (repres.) repression of protein production; (dead.) deadenylation; (decay) degradation of the mRNA body. (B) Interactions between HA-CUP wild type or mutants and endogenous eIF4E. Proteins were immunoprecipitated from cell lysates using a monoclonal anti-HA antibody. An HA-tagged version of maltose-binding protein (MBP) served as negative control. Inputs (1%) and immunoprecipitates (10%) were analyzed by Western blotting using anti-HA and anti-eIF4E antibodies. (C–F) The activity of CUP mutants relative to wild type was tested in tethering assays using the F-Luc-5BoxB reporter and analyzed as described in Figure 1. The mean values ± standard deviations from three independent experiments are shown. (G) Wild-type CUP and mutants were expressed at comparable levels. R-Luc-V5 served as a transfection control.