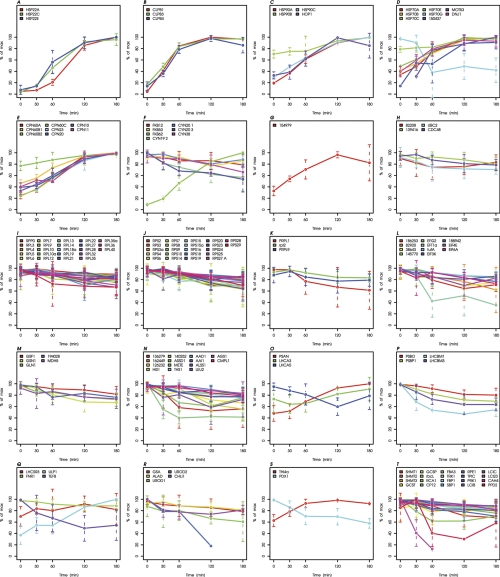
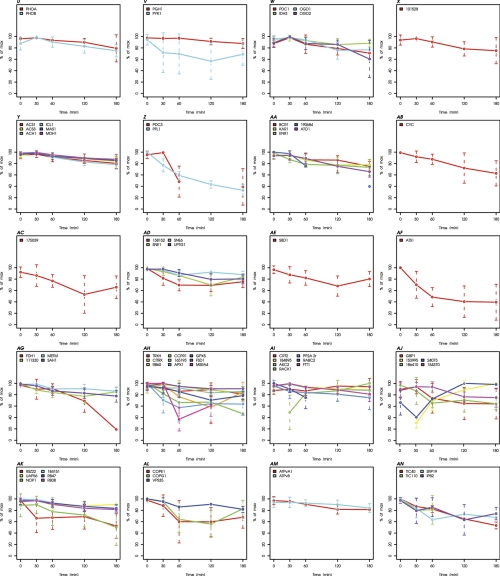
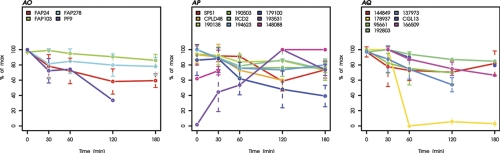
Fig. 4.
Kinetics of proteins exhibiting significant changes during heat stress. Kinetics of all 244 proteins significantly changing during heat stress are plotted bin-wise as percent of the maximal value in the time course. Values and standard deviations derive from three biological and two technical replicates (listed in supplemental Table S3). A, Small heat shock proteins. B, HSP100s. C, HSP90 and co-chaperones. D, HSP70 and co-chaperones. E, HSP60s. F, FKBPs/Cyclophilins. G, Protein folding and abiotic stress. H, Proteases/Peptidases. I, Protein biosynthesis (LSU cytosol). J, Protein biosynthesis (SSU cytosol). K, Protein biosynthesis (LSU plastid). L, Protein biosynthesis (not ribosome). M, N-metabolism. N, Amino acid metabolism. (O) Proteins associated with PS I. P, Proteins associated with PS II. Q, Other proteins associated with light reactions. R, Porphyrin biogenesis. S, Vitamin biosynthesis. T, Proteins of photosynthetic and photorespiratory carbon metabolism. U, Starch synthesis and degradation. V, Glycolysis and gluconeogenesis. W, TCA cycle. X, Oxidative pentose phosphate pathway. Y, Acetate metabolism/glyoxylate cycle. Z, Fermentation. AA, Glycerolipid metabolism. AB, Mitochondrial electron transfer chain and ATP synthase. AC, Nucleotide metabolism. AD, Sugar nucleotide metabolism. AE, Metal handling. AF, S-assimilation. AG, C1-metabolism. AH, Redox regulation/oxidative stress response. AI, Signal transduction. AJ, DNA binding/transcription/chromatin remodeling. AK, RNA processing/binding. AL, Vesicle transport. AM, Pumps and transporters. AN, Protein sorting. AO, Flagellar and basal body proteins. AP, Miscellaneous. AQ, Proteins without functional annotation.