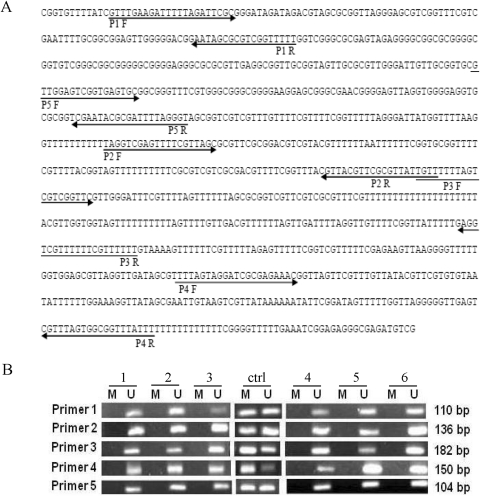
Fig. 6.
A, NT5C2 promoter with CpG sites. Bisulfite-modified genomic DNA sequence of the CpG region of the human NT5C2 gene. The position of the primers that were used for methylation-specific PCR after bisulfite treatment are indicated by forward and reverse arrows for primer 1 to primer 5 covering five different regions of the CpG island. The bisulfite treatment converted the unmethylated cytosine to uracil, which is complementary to adenosine. B, methylation analysis of five CpG sites in the NT5C2 promoter. Representative agarose gels showing methylation patterns in sensitive and resistant HapMap (CEU) cell lines for the NT5C2 primer1 to primer 5. M, band for methylated primers; U, band for unmethylated primers. The positive control (ctrl) methylated and unmethylated bands was used from EpiTect PCR Control DNA Set.