FIGURE 1.
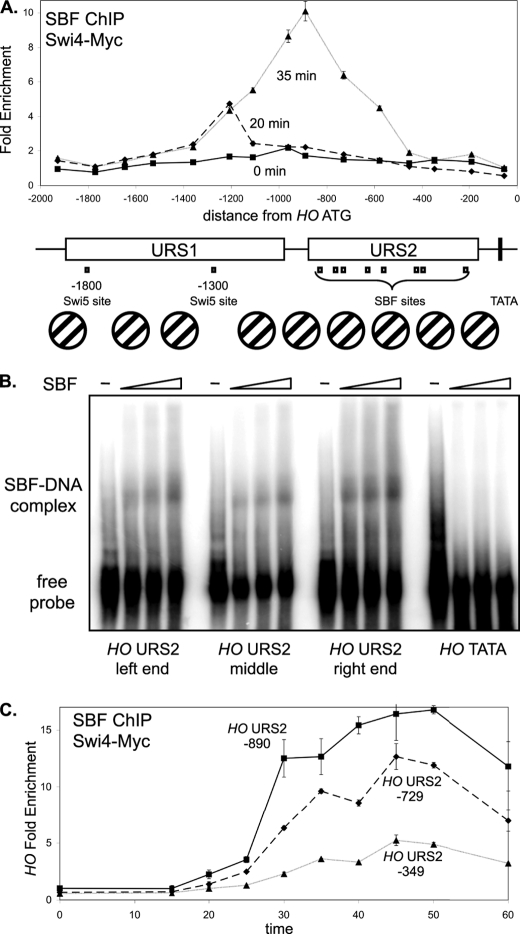
SBF binding is strongest to sites at the left end of URS2. A, DY12794 cells (GALp::CDC20 SWI4-Myc) with a GALp::CDC20 allele were synchronized in mitosis by removing galactose, followed by release by addition of galactose (t = 0). The CDC20 arrest is at the G2/M transition, and HO expression at 40 min following release corresponds to late G1 phase. SBF binding was measured by Swi4-Myc ChIP using samples taken at t = 0, 20, and 35 min following the release. ChIP samples were analyzed with 15 sets of PCR primers across the HO promoter, with an average PCR product size of 208 bp. URS1-, URS2-, Swi5- and SBF-binding sites are shown for the HO promoter, where the ATG represents +1 and the transcription start site is at −20. Nucleosome positions along the HO promoter, determined by micrococcal nuclease mapping and by H3 ChIP (3), are shown. B, electrophoretic mobility shift assays examine in vitro binding of recombinant SBF to sites at the left (−857), middle (−461) and right (−246) parts of URS2, and the TATA region as a negative control. C, DY12794 cells (GALp::CDC20 SWI4-Myc) were synchronized and SBF binding by Swi4-Myc ChIP during the time course to three regions of HO URS2 at −890, −729, and −349, where the numbers refer to the center of the amplified region.