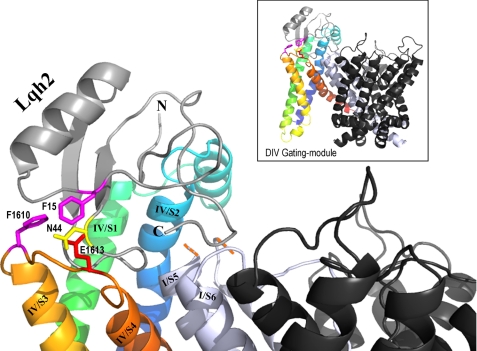
FIGURE 5.
Model of Lqh2 interaction with rNav1.2 resting state. The external loops DIV/S1-S2 and S3-S4 of rNav1.2a were constructed on the structural model of Kv1.2 in its resting state (43) using the Swiss-PdbViewer. The internal loops and the gating modules of DI, DII, and DIII were removed. Shown in black are the remaining pore modules of DII, DIII, and DIV, whereas the DI pore module is shown in light gray. Due to the substantial difference in size, the DI/S5-SS1 external loop is omitted (indicated by the orange dashes). DIV/S1 is shown in green, DIV/S2 is shown in blue, DIV/S3 is shown in light orange, and DIV/S4 is shown in dark orange. Lqh2 modeling relied on its close resemblance to Aah2 (29; Protein Data Bank code 1AHO). Phe-15 and Asn-44 are bioactive residues of Lqh2 that are in close proximity to Phe-1610 and Glu-1613 of the channel, respectively (colored sticks according to their chemical nature). Docking of Lqh2 core domain at DIV gating module was performed using DockingServer. Although the residues of the toxin NC domain that may interact with residues at DI/S5-SS1 and DIV/S1-S2 have not been clarified yet, further modeling was performed manually to show this proximity, while avoiding side chain clashes. The final figure was drawn using PyMOL.