Background: GPBP-1 is a non-conventional kinase that regulates glomerular basement membrane collagen organization.
Results: GPBP-1 targets GIP130, a new myosin-binding protein, and regulates myofibrillogenesis in cultured myoblasts.
Conclusion: GPBP-1 regulates myofibril formation.
Significance: GPBP-1 is a kinase for structural protein organization at both intracellular and extracellular compartments.
Keywords: Cell Differentiation, Cytoskeleton, Differentiation, Gene Knockout, Myosin, Protein-Protein Interactions, Skeletal Muscle, CERT, FILIP1L, GPBP
Abstract
Goodpasture antigen-binding protein-1 (GPBP-1) is an exportable non-conventional Ser/Thr kinase that regulates glomerular basement membrane collagen organization. Here we provide evidence that GPBP-1 accumulates in the cytoplasm of differentiating mouse myoblasts prior to myosin synthesis. Myoblasts deficient in GPBP-1 display defective myofibril formation, whereas myofibrils assemble with enhanced efficiency in those overexpressing GPBP-1. We also show that GPBP-1 targets the previously unidentified GIP130 (GPBP-interacting protein of 130 kDa), which binds to myosin and promotes its myofibrillar assembly. This report reveals that GPBP-1 directs myofibril formation, an observation that expands its reported role in supramolecular organization of structural proteins to the intracellular compartment.
Introduction
The human COL4A3BP gene expresses at least three polypeptides, including canonical GPBP-1 (previously GPBP)3 of 77 kDa; GPBP-2 (previously GPBPΔ26/CERT), which lacks the 26-residue serine-rich region encoded by exon 11 due to alternative exon splicing; and GPBP-3 (previously 91-kDa GPBP), a variant containing 83 additional residues derived from alternative mRNA translation initiation. GPBP-1 is secreted in a soluble form, GPBP-2 localizes in the cytosol, and GPBP-3 is associated with cellular membranes (1).
GPBP-1 is a non-conventional Ser/Thr kinase that binds and phosphorylates noncollagenous domain-1 of the α3 chain of type IV collagen (2), the target of autoantibodies in Goodpasture autoimmune disorder also known as the Goodpasture (auto)antigen (3). Noncollagenous domain-1 directs basement membrane collagen (type IV) molecular and supramolecular organization (4). Increased GPBP-1 expression has been shown to induce glomerular basement membrane collagen disorganization (5). These and other lines of evidence suggest that GPBP-1 is involved in regulating folding and assembly of glomerular basement membrane collagen in the secretory and extracellular compartments (1, 5).
Approaches directed at elucidating the biological activity of GPBP-2 include pioneer studies suggesting that GPBP-2 is less active than GPBP-1 (6) and subsequent studies suggesting that GPBP-2 serves as a ceramide transporter between the endoplasmic reticulum (ER) and the Golgi apparatus for regulation of protein secretion (7, 8). Finally, more recent studies have implicated GPBP-3 in promoting GPBP-1 exportation (1). Collectively, the data suggest that COL4A3BP products orchestrate a biological program that regulates protein folding and secretion (1, 5, 8).
The biological functions of the COL4A3BP orthologs in Drosophila melanogaster, Danio rerio, and Mus musculus have been investigated by chemical mutagenesis, knockdown, and knock-out approaches, respectively (9, 10, 11). In flies, the col4a3bp gene (GenBankTM accession number CG7207) encodes a single polypeptide dmGPBP-1 (GenBankTM accession number NP_648199), whose absence causes increased oxidative stress, accelerated aging, and reduced life span (9). In zebrafish, col4a3bp generates both drGPBP-1 and -2 (GenBankTM accession numbers EU000165 and EU000166), and drGPBP-1, but not drGPBP-2, exerts an antiapoptotic activity that is critical for normal muscle and brain development and embryo survival (10). Mouse Col4a3bp also encodes mGPBP-1 and -2 (GenBankTM accession numbers AF232932 and AF232934); however, Col4a3bp-null mice were not viable due to defective heart development associated with mitochondria degeneration, ER alterations, chronic ER stress, and defective cell proliferation (11).
Because COL4A3BP is mostly expressed in striated muscle (2) and an orthologous gene is required for muscular development in lower vertebrates (10), we investigated the role of mouse Col4a3bp in myogenesis. Our results reveal that Col4a3bp is critical for myogenesis and that GPBP-1 mediates myofibril myosin assembly through the targeting of the previously unrecognized GIP130 (GPBP-interacting protein of 130 kDa). Collectively, our data reveal that GPBP-1 regulates supramolecular organization of structural proteins at both extracellular (i.e. type IV collagen) and intracellular (i.e. myosin) compartments.
EXPERIMENTAL PROCEDURES
Mice
To obtain gpbp-1−/− mice (Neo+/LoxP+), embryonic stem cells obtained as indicated in the supplemental material were injected into C57BL/6 blastocysts to generate chimeric mice containing the target sequence. Chimeric mice were crossed with C57BL/6 mice to yield heterozygous (Neo+/LoxP+) individuals, which were inbred to yield homozygous (Neo+/LoxP+) mice. Finally, homozygous (Neo+/LoxP+) mice were subsequently crossed with transgenic BALB/c-Tg(CMV-cre)1Cgn/J mice (The Jackson Laboratory), ubiquitously and constitutively expressing Cre recombinase. The resulting offspring were genotyped for exon 11 knock-out identification (supplemental Fig. S1). Homozygous Col4a3bp exon 11 knock-out mice (gpbp-1−/−) and their littermates lacking genetic deletion (gpbp-1+/+) were maintained as an inbred colony of mixed genetic background. Previously reported Tg-hGPBP-1 mice (5) were back-crossed with C57BL/6 mice until they reached 99.2% C57BL/6 genetic background. Tg-hGPBP-1 mice and littermates containing no genetic modification (WT) used in the present studies contained 96.3–99.2% C57BL/6 genetic background as determined by genetic characterization of 222 independent genetic markers using speed congenic procedures (Bionostra, Madrid, Spain). For some studies, inbred C57BL/6 mice were used as a source of striated muscle material for biochemical and morphological studies. All of the procedures were performed according to institutional guidelines for the use of animals in experimentation. The age of the mice used for the studies varied between 2 and 10 months. The age of genetically unmodified (control) and genetically modified (gpbp−/− and Tg-hGPBP-1) mice was similar for each individual procedure.
Cell Culture
Mouse C2C12 cells and immortalized myoblasts were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% fetal calf serum (FCS) for normal propagation. Myogenic differentiation was obtained by culturing cells in DMEM supplemented with 2% horse serum. Human embryonic kidney (HEK) 293 cells were cultured in DMEM supplemented with 10% FCS. All media contained 100 units/ml penicillin/streptomycin.
Antibodies
Monoclonal antibodies were produced essentially as previously reported (12) using GST-FLAG-GIP90 (GIP90 is a shorter isoform of GIP130; see supplemental material for a full explanation) as immunogen. We obtained antibodies reacting with both recombinant (r) GIP130 and rFILIP1L-102 (mAb8GIP), and only the clones expressing antibodies that recognized rGIP130 and not rFILIP1L-102 were used in the present studies (mAb3GIP). The production and characterization of mouse mAb 14, mAb e26, mAb N27, and rabbit polyclonal anti-GPBP antibodies have been reported (1, 2, 13). Anti-myosin heavy chain (MyoHC) mouse monoclonal (Sigma) and rabbit polyclonal (Abcam) antibodies were used for myosin detection. Anti-GPBP chicken polyclonal and anti-Pax rabbit polyclonal antibodies were from Abcam. Mouse mAbs specific for caveolin 3, M-cadherin, MyoD, and myogenin were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); those for tubulin and FLAG tag were from Sigma; and those for GAPDH were a gift of Erwin Knecht. Akt and phospho-Thr308-Akt were detected with rabbit polyclonal and monoclonal antibodies, respectively (Cell Signaling Technology).
Derivation of Myoblast Cell Lines from Mouse Skeletal Muscle Satellite Cells
Three-month-old gpbp-1+/+ (n = 2) and gpbp-1−/− (n = 2) littermate male mice were used to derive independent myoblast cell lines. Likewise, 2-month-old Tg-hGPBP-1 (n = 2) and WT (n = 2) male littermate mice were used for similar purposes. Briefly, satellite cells were isolated from hind limb muscles of individual mice and cultured as described previously (14) until spontaneous immortalization was achieved. Immortalized cells were subsequently cultured for propagation and myogenic differentiation.
Production of Other Cell Lines for Recombinant Protein Expression
One million HEK 293 cells were transfected with 10 μg of pTRE2hyg-n4′ construct expressing GPBP-1 under the control of a doxycyclin-inducible promoter. Transfected cells were selected with 200 μg/ml hygromycin and further maintained with 100 μg/ml hygromycin in culture medium. Half a million C2C12 mouse cells were transfected with 10 μg of pRK5-c-Myc-GIP130-EGFP and 2 μg of pTRE2hyg. Transfected cells were selected with 200 μg/ml hygromycin and further maintained with 100 μg/ml hygromycin in suitable culture medium. Transfections were performed either with the Profection Mammalian Transfection System-Calcium Phosphate (Promega) or with Lipofectamine (Invitrogen).
Growth Rate Determination of Cell Lines
Thirty thousand cells of individual gpbp-1+/+ (n = 2) and gpbp-1−/− (n = 2) myoblast cell lines were seeded on 6-well plates and cultured in growth medium. Cells were trypsinized and counted with trypan blue stain (Cambrex Bio Science) in a hemocytometer at 24, 48, and 72 h after seeding.
Myofibril Isolation
Myofibrils were isolated from mouse hind legs essentially as previously described (15).
Morphological Studies
Phase-contrast, conventional immunofluorescence, and confocal microscopy; histochemistry; immunohistochemistry; and electron microscopy (EM) studies were performed using methods described in the supplemental material.
Protein Extraction, Cell Fractioning, Immunoprecipitation, and Pull-down Assays
Mouse quadriceps were ground in a mortar with liquid nitrogen on a dry ice bed and homogenized in 50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 1% Triton X-100, 0.1% SDS, 10 μg/ml leupeptin, 1 mm PMSF with a Polytron homogenizer (Omni International). Homogenates were maintained for 16 h at 4 °C with gentle rocking and cleared by centrifugation at 16,000 × g for 5 min at 4 °C. When indicated, isolated myofibrils were suspended in the same homogenization buffer and treated similarly. To extract cell cultures for detection of GPBPs, GIP130, and MyoHC as well as for immunoprecipitation, cells were lysed with 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 10 μg/ml leupeptin, 1 mm PMSF. For detection of myogenin, MyoD, Pax7, M-cadherin, and caveolin-3, cells were lysed with 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.1% SDS, 1% sodium deoxycolate, 1% Triton X-100, 10 μg/ml leupeptin, 1 mm PMSF. For detection of Akt and phospho-Thr308-Akt, cultures were lysed in 50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 1% Tween 20, 0.2% Nonidet P-40, 50 mm β-sodium glycerophosphate, 0.2 mm sodium orthovanadate, 10 μg/ml leupeptin, 1 mm PMSF. All cell protein extractions were performed for 30 min at 4 °C, and homogenates were cleared as above. Protein concentration was determined in supernatants using the Bio-Rad protein assay. All extracts were stored at −20 °C until use. Cell fractioning was performed essentially as described previously (1).
FLAG-GPBP-1 was precipitated from either cultured myoblasts or myofibril extracts of Tg-hGPBP-1 mice with anti-FLAG affinity resin (Sigma) according to the manufacturer's recommendations. Prior to immunoprecipitation, myofibril extracts were brought to 150 mm NaCl with 50 mm Tris-HCl, pH 7.5.
For co-immunoprecipitation of mGPBP-1 and mGIP130, protein extracts from diaphragm (500 μg) were brought to 150 mm NaCl as above, precleared with 50 μl of protein A-Sepharose (2 h), and incubated with or without 5 μg of mAb N27 (16 h), and then 50 μl of protein A-Sepharose was added (2 h), and beads were collected by centrifugation. For co-immunoprecipitation of recombinant proteins, HEK 293 cells expressing rGPBP-1 in a doxycyclin-inducible fashion were transfected either with pRK5-c-Myc-GIP130 or with pRK5-c-Myc and cultured for 24 h, and rGPBP-1 expression was induced with 1 μg/ml doxycyclin for an additional 24 h. Cells were lysed, and protein extracts were similarly precleared and extracted with anti-c-Myc-agarose affinity gel (Sigma).
For pull-down assays, 1 mg of mouse myofibrils were centrifuged at 16,000 × g for 5 min, and the pellet was dispersed by sonication in 50 mm Tris-HCl, 150 mm NaCl, 8 m urea, pH 7.5. Five hundred micrograms of urea-solubilized myofibrillar material were dialyzed against Tris-buffered saline in the presence or absence of 2 μg of FLAG-GIP130 (16 h) and centrifuged at 1000 × g (3 min) for clearing purposes, and supernatants were extracted with anti-FLAG affinity resin. In co-immunoprecipitation and pull-down procedures, loaded beads were extensively rinsed with 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100 (three times for 15 min each), and bound material was eluted with SDS-PAGE sample buffer. All immunoprecipitation procedures were performed at 4 °C with gentle rocking.
Phosphorylation and Dephosphorylation Assays, Western Blot, and Far Western
These procedures were performed essentially as described previously (1, 2, 6).
Densitometry
When indicated, the amount of reactive polypeptides in Western blots was estimated with AlphaImager software (Cell Biosciences).
Expression and Purification of Recombinant Proteins
These procedures were performed following standard procedures described in the supplemental material.
RNA Interference Assays
For these studies, we designed an shRNA approach using pSi-GPBP/GPBPΔ26-2, a construct previously reported to target human GPBP mRNAs (1). Despite the existence of three mismatches between human and mouse mRNA targeted sequence, this construct efficiently suppressed mGPBP expression in individual C2C12 clones. Specifically, 0.5 × 106 C2C12 cells were seeded and cultured overnight at 37 °C and the following day were transfected by standard calcium phosphate procedures with 10 μg of pSi-GPBP/GPBPΔ26-2 and 2 μg of pEGFP-N1 (Clontech). Ten days after, cells positive for green fluorescence were sorted on a 96-well culture plate with a MoFlo Cell Sorter (Beckman Coulter), and further cultured. Individual clones were assessed for Col4a3bp expression by Western blot procedures using mAb 14 or subjected to differentiation.
A similar shRNA approach was followed for silencing mGIP130. In this case, C2C12 cells were transfected with 10 μg of pSi-U6-mGIP-2 (see supplemental material) and 2 μg of pEGFP-N1 and similarly processed. The mGIP130 expression levels of individual clones were assessed by quantitative RT-PCR. The pSilencer 2.1 U6-hygro vector (Ambion) was used to develop control clones.
Cytoskeleton Extraction Assays
Cytoskeleton extraction assays were performed essentially as described previously (16). For other experimental procedures, see the supplemental material.
RESULTS
Col4a3bp Expression Regulates C2C12 Myoblast Differentiation
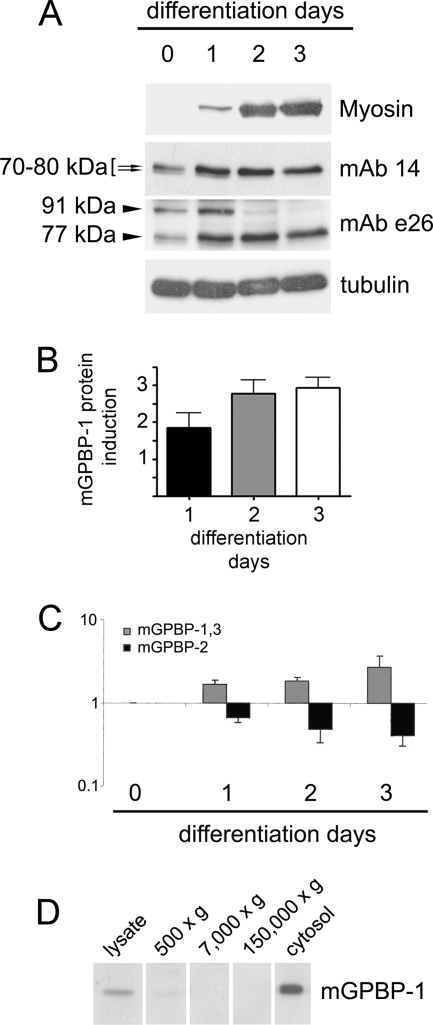
To investigate the role of Col4a3bp in striated muscle development, we first analyzed its expression in C2C12 cultures, a well established model of myogenesis (Fig. 1). Upon serum deprivation, C2C12 myoblasts fuse into large multinucleated myotubes, which express skeletal muscle myosin and form myofibrils, the contractile machinery of muscles (17). We subjected C2C12 cultures to differentiation and assessed myogenesis by monitoring the expression of fast MyoHC. In undifferentiated cells, mAb 14, recognizing human intracellular GPBP-1 and -2 but not GPBP-3 (1, 2), detected two closely migrating polypeptides of 70–80 kDa that represented mGPBP-1 and -2 and that were oppositely regulated during differentiation (Fig. 1A, arrows). To assess the expression of mGPBP-1 and -3, we used mAb e26, which recognizes only these isoforms in human cells (1). We found that serum deprivation induced the counterregulation of two reactive polypeptides of 77 and 91 kDa representing mGPBP-1 and -3, respectively (Fig. 1A, mAb e26). These observations suggested that differentiation of C2C12 cells was associated with down-regulation of mGPBP-2 and -3 and concomitant up-regulation of mGPBP-1 expressions. Increased expression of GPBP-1 during differentiation was confirmed by densitometry (Fig. 1B) and quantitative RT-PCR, which showed an opposite regulation of exon 11-containing (mostly GPBP-1) and exon 11-devoid (GPBP-2) mRNAs during C2C12 myogenesis (Fig. 1C). Furthermore, Western blot analysis of subcellular fractions of a 3-day differentiated myoblast culture demonstrated mGPBP-1 to be mainly expressed in the cytosol (Fig. 1D).
FIGURE 1.
Col4a3bp expression in differentiating C2C12 myoblasts. A, C2C12 cells were serum-deprived, and at the indicated days, 50 μg of cell lysates were subjected to Western blot analysis with antibodies targeting MyoHC (Myosin), GPBP-1 and -2 (mAb 14), GPBP-1 and -3 (mAb e26), and tubulin. The bracketed arrows and arrowheads denote the position of GPBP polypeptides of the indicated molecular mass. B, the protein expression of mGPBP-1 in C2C12 differentiation assays (n = 2) was assessed by Western blot and quantified by densitometry with respect to tubulin expression. C, the RNA extracted from the cells in A was used for quantitative RT-PCR to determine the relative expression of the indicated mRNAs (bars). B and C, values are in -fold expression with respect to undifferentiated cells (day 0), which was set at 1. D, 3-day differentiated C2C12 cells were subjected to subcellular fractionation, and 15 μg of each of the indicated fractions were analyzed by Western blot for specific mGPBP-1 detection with mAb e26. Error bars, S.D.
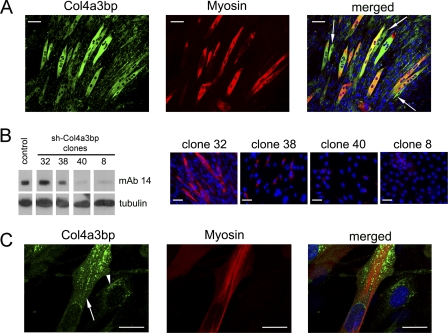
To further study the role of intracellular accumulation of mGPBP-1 in myogenesis, 3-day differentiated C2C12 cultures were subjected to confocal microscopy analysis. A number of differentiating cells showed high levels of intracellular mGPBP-1; however, not all cells expressing intracellular mGPBP-1 at high levels displayed myosin expression (arrows), but all myosin-expressing cells showed elevated intracellular mGPBP-1 expression (Fig. 2A). These observations suggested that intracellular accumulation of mGPBP-1 preceded the onset of myosin synthesis. To test whether myogenesis was dependent on Col4a3bp expression in C2C12 cells, we designed a strategy based on shRNA interference and produced cell lines expressing Col4a3bp at different levels (Fig. 2B). We found a correlation between expression of Col4a3bp and myosin synthesis in differentiating cells and thus moderate reductions in Col4a3bp expression (clone 38) associated with diminished levels of myosin, whereas extensive silencing of Col4a3bp (clones 40 and 8) prevented differentiation of C2C12 cells. Confocal microscopy analysis of differentiating C2C12 cells showed mGPBP-1 aggregates in the juxta- and perinuclear region in less differentiated cells (arrowhead) and decorating the nuclear envelope, accumulating away from the nucleus and forming rows between nascent myofibrils in more differentiated cells (arrow) (Fig. 2C). Collectively, these observations suggested that intracellular accumulation of mGPBP-1 was critical for myogenesis.
FIGURE 2.
Col4a3bp expression regulates myogenesis in C2C12 myoblasts. A, C2C12 cells were subjected to a 3-day differentiation and analyzed by confocal microscopy using rabbit polyclonal antibodies specific for Col4a3bp products and a mouse mAb for MyoHC detection. The white arrows indicate cells with increased intracellular expression of mGPBP-1 in which MyoHC could not be detected. Bars, 60 μm. B, C2C12 cells were transfected with construct pSi-GPBP/GPBPΔ26–2, and the indicated clones were analyzed either for Col4a3bp (mAb 14) or tubulin expression using Western blot procedures (left) or subjected to a 3-day differentiation and MyoHC expression analyzed as in A using conventional immunofluorescence microscopy (right). Bars, 30 μm. C, C2C12 cells were differentiated for 3 days, and mGPBP-1 and MyoHC expression was analyzed as in A. The arrow and arrowhead pinpoint mGPBP-1 aggregates in more and less differentiated cells, respectively. Bars, 20 μm. In this and following figures, nuclei are DAPI-stained and displayed in blue.
Deficiency of mGPBP-1 Impairs Myofibril Organization in Differentiating Myoblasts
It has been reported that complete inactivation of Col4a3bp causes embryonic lethality (11). Therefore, to investigate the role of mGPBP-1 in myogenesis, we adopted the alternative strategy of deleting exon 11 of Col4a3bp to abrogate the expression of mGPBP-1 and -3 while preserving that of mGPBP-2 (supplemental Fig. S1). Given the observations that mGPBP-3 expression is down-regulated in myogenesis, our model permitted us to explore the specific role of mGPBP-1 in this process. In contrast to mice bearing complete inactivation of Col4a3bp, our gpbp-1−/− mice developed normally and displayed regular fertility. No differences in body weight and size were observed between gpbp-1+/+ and gpbp-1−/− mice matched by sex and age (3–12 months) (data not shown).
We obtained myoblast cultures from gpbp-1+/+ and gpbp-1−/− mice and characterized them by Western blot analysis with mAb e26 (Fig. 3A). As expected, gpbp-1+/+ myoblasts expressed mainly mGPBP-1 and traces of mGPBP-3, whereas in gpbp-1−/− myoblasts, neither isoform was detected. When the extracts were analyzed with mAb 14, both mGPBP-1 and -2 were detected in gpbp-1+/+ cell lysates, and only mGPBP-2 was expressed in the gpbp-1−/− counterparts, although more abundantly than in gpbp-1+/+ lysates. These observations were confirmed by conventional and quantitative RT-PCR procedures (Fig. 3B).
FIGURE 3.
Myoblast cell lines derived from gpbp-1−/− mice show impaired differentiation. A, 50 μg of lysates from gpbp-1+/+ (+/+) or gpbp-1−/− (−/−) myoblasts were either analyzed by Western blot for mGPBP-1 and -3 (mAb e26) and tubulin detection or dephosphorylated with λ-phosphatase and similarly analyzed to detect mGPBP-1 and -2 (mAb 14). B, the cells in A were subjected to RNA extraction, RT, and conventional (left) or quantitative (right) PCR to assess levels of the indicated mRNAs. The products of the conventional PCR were analyzed by agarose gel electrophoresis and stained with ethidium bromide. Numbers and bars indicate the size (bp) and position of the standard DNA markers, and the arrowheads denote the position of the indicated cDNA products. The plot reflects the relative expression of the indicated mRNA as -fold level of gpbp-1−/− (−/−) with respect to gpbp-1+/+ (+/+), which was set at 1. C, myoblasts from gpbp-1+/+ (+/+) and gpbp-1−/− (−/−) mice were differentiated for 5 days and analyzed by phase-contrast microscopy. Original magnification was ×100. D, the cultures in C were stained with antibodies for MyoHC (green), and individual representative cells were analyzed by confocal microscopy. Inset, characteristic striation of myofibrils assembled in differentiated gpbp-1+/+ (+/+) cells. Bars, 20 μm. E, the cells in C were analyzed by EM to visualize myofibrillar material. Note the contrast between myofibrillar material in gpbp-1+/+ myoblasts, which displayed well developed sarcomeric organization and myofibrillar material in gpbp-1−/− myoblasts lacking sarcomeric organization. F, micrograph showing a cytoplasmic body characteristic of gpbp-1−/− myoblast cultures (left) and a magnified cytoplasmic body displaying radial distribution of myofibrillar material (right). G, 50 μg of lysates from undifferentiated (U) or 5-day differentiated (D) gpbp-1+/+ (+/+) or gpbp-1−/− (−/−) myoblasts were subjected to Western blot analysis with antibodies against the indicated proteins. H, 30,000 myoblasts from gpbp-1+/+ (+/+) (n = 2) and gpbp-1−/− (−/−) (n = 2) mice were cultured in growth medium, and cells were quantified at days 1, 2, and 3 after seeding. Division rates were estimated and represented as means ± S.E. (error bars) of -fold growth at the indicated times after day 1. Differences in growth rate between +/+ and −/− cultures lacked statistical significance.
To further investigate the role of mGPBP-1 in myogenesis, gpbp-1+/+ and gpbp-1−/− myoblasts cultures were serum-deprived for 5 days, and differentiation was monitored in parallel using morphological techniques. Interestingly, gpbp-1−/− cultures showed remarkable defective cell fusion and myotube formation (Fig. 3C). Furthermore, myosin formed tight bundles characteristic of myofibrils in gpbp-1+/+ cultures, whereas it appeared diffused throughout the cell cytoplasm, accumulating in loose fibrils without sarcomere organization in gpbp-1−/− cultures (Fig. 3D). These findings were further confirmed by EM analysis, which revealed well organized nascent myofibrils in differentiating gpbp-1+/+ cells and non-sarcomeric fibrils in gpbp-1−/− differentiating myoblasts (Fig. 3E). Moreover, in gpbp-1−/− differentiating myoblasts, we observed aggregates of fibrillar material (cytoplasmic bodies), often without defined organization (Fig. 3F, left) and occasionally with a radial arrangement (Fig. 3F, right). To investigate defective myogenesis in gpbp-1−/− cultures, we analyzed in parallel with gpbp-1+/+ the expression of critical myoblast differentiation biomarkers, including MyoHC; myogenic transcription factors MyoD (18), myogenin (19), and Pax7 (20); AKT kinases (21, 22) and their phosphorylated Thr308 counterparts (23); and proteins mediating cell fusion, caveolin-3 (24) and M-cadherin (25) (Fig. 3G). Also, we estimated the cell division rates (Fig. 3H). Other than the lack of GPBP-1 expression (Fig. 3A), we did not find significant biological differences between gpbp-1−/− and gpbp-1+/+ cultures to account for defective differentiation of gpbp-1−/− cultures.
Human Recombinant GPBP-1 Promotes Myofibril Assembly in Cultured Mouse Myoblasts
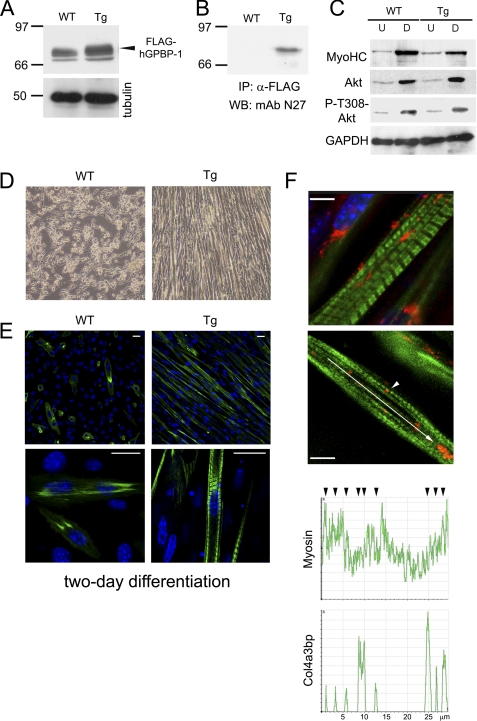
To further study the relevance of GPBP-1 in myogenesis, C57BL/6 transgenic mice expressing FLAG-tagged human GPBP-1 (Tg-hGPBP-1) and C57BL/6 (WT) mice were used to derive myoblast cell lines. Western blot analysis of cell extracts using mAb N27, an antibody reactive with both GPBP-1 and -2, revealed that Tg-hGPBP-1 myoblasts expressed higher levels of 70–80-kDa reactive polypeptides than WT myoblasts (Fig. 4A, arrowhead). Immunoprecipitation of the cell lysates with anti-FLAG antibodies confirmed the presence of FLAG-hGPBP-1 only in Tg-hGPBP-1 myoblasts (Fig. 4B). When these cultures were subjected to differentiation, no major differences were observed in the expression of MyoHC, Akt, or phospho-Thr308-Akt (Fig. 4C). However, myoblast alignment and fusion as well as myofibril formation were more efficient in Tg-hGPBP-1 cultures than in their WT counterparts. Thus, Tg-hGPBP-1 cultures showed abundant myotube formation after a 2-day differentiation, whereas in parallel cultured WT myoblasts showed a more limited degree of differentiation (Fig. 4, D and E). Finally, confocal microscopy analysis revealed that GPBP-1 distribution in Tg-hGPBP-1 cultures was similar to that of C2C12 myoblasts and thus formed large aggregates, which were mainly juxta- and perinuclear in less differentiated cells and intermyofibrillar in more differentiated myoblasts (Fig. 4F, top image). In the latter case, GPBP-1 aggregates were also observed in a quasiregular distribution along myofibrils acquiring striated organization (arrow) and associated with myosin at band A (arrowhead) in more developed myofibrillar structures (Fig. 4F, bottom image and graphs).
FIGURE 4.
Myoblast cell lines derived from Tg-hGPBP-1 mice show accelerated differentiation. A, lysates of undifferentiated myoblasts derived from wild type (WT) or Tg-hGPBP-1 (Tg) mice were analyzed by Western blot with specific antibodies for GPBP (mAb N27) or for tubulin, used as loading control. The arrowhead indicates the position of reactive polypeptide present only in Tg-hGPBP-1 lysates representing FLAG-tagged GPBP-1. B, the lysates in A were subjected to immunoprecipitation (IP) and Western blot analysis of precipitates (WB) using the indicated antibodies. Numbers and bars in A and B denote the size (kDa) and position of individual molecular mass markers. C, lysates (50 μg) of undifferentiated (U) and 4-day differentiated (D) WT or Tg-hGPBP-1 myoblasts were analyzed by Western blot using antibodies specific for the indicated proteins. D, the cells in A were differentiated for 2 days and analyzed by phase-contrast microscopy. Original magnification was ×100. E, representative confocal microscopy images of the cultures in D stained with antibodies for MyoHC visualization (green). Bars, 20 μm. F, representative confocal microscopy images of a differentiated Tg-hGPBP-1 culture stained with MyoHC-specific (green) and GPBP-specific (red) antibodies. The arrowhead denotes GPBP-1 intimately associated with an A band in a striated myofibril. Bars, 5 μm. The graphs represent the distribution of fluorescence intensity in the region indicated by the arrow in a nascent myofibril. The arrowheads denote co-localization of the indicated proteins.
GPBP-1 Interacts and Phosphorylates Previously Unrecognized Myofibrillar Component GIP130
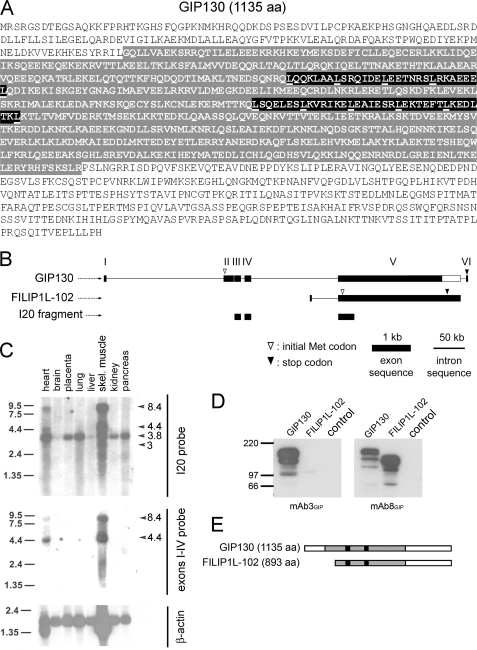
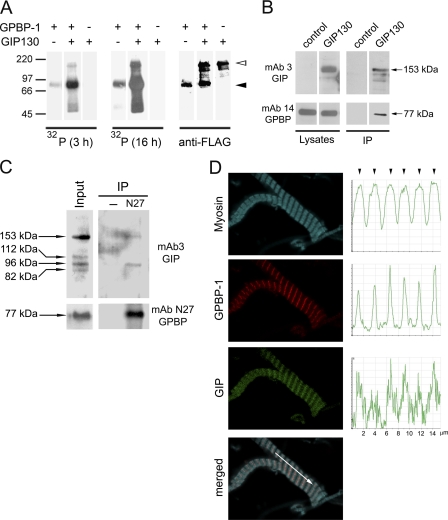
To identify downstream effectors of GPBP-1 involved in myogenesis, we screened a yeast two-hybrid human cDNA striated muscle library using GPBP-1 as bait and isolated I20, a truncated cDNA that comprised previously unrecognized exon sequence but also sequence present in FILIP1L mRNA (previously known as DOC1; GenBankTM accession number BC027860) (see supplemental material). Using standard hybridization procedures, we identified a cDNA predicting GIP130 (GPBP-interacting protein of 130 kDa) (GenBankTM accession number AF514867) (Fig. 5A). GIP130 is a new isoform of the FILIP1L protein family, which is generated by N terminus extension of the open reading frame of FILIP1L mRNA, predicting a 102-kDa polypeptide here named FILIP1L-102 (Fig. 5B). FILIP1L was widely expressed in human tissues displaying preferential expression in striated muscle; however, GIP130 displayed a more restricted tissue distribution and was virtually only expressed in striated muscle (Fig. 5C). We have characterized four less abundant isoforms of GIP130 (GenBankTM accession numbers AF329092, AF514868, AF514869, and AY642382) that unlike FILIP1L-102 were also found to interact with GPBP-1 and to be preferentially expressed in striated muscle.4 To investigate the biological significance of GIP130, we produced rGIP130 and rFILIP1L-102 and developed GIP130-specific monoclonal antibodies (mAb3GIP) as well as monoclonal antibodies recognizing both isoforms (mAb8GIP) (Fig. 5D). GIP130 was expressed as a major 153-kDa product that contained a large coiled-coiled domain with two leucine zippers, suggesting that it was a structural polypeptide preferentially expressed in striated muscle (Fig. 5E).
FIGURE 5.
GIP130 is a FILIP1L isoform having features of a structural protein that is preferentially expressed in striated muscle. A, the primary structure of GIP130 is shown in one-letter code. The predicted coiled-coiled region and canonical leucine zippers are highlighted with gray and black backgrounds, respectively. B, shown are the exon-intron structures of pre-mRNA of GIP130 (GenBankTM accession number AF514867) and FILIP1L-102 (GenBankTM accession BC027860) transcribed from FILIP1L at 3q12.1 and exon sequence of I20. C, 32P-labeled cDNAs representing both GIP130 and FILIP1L-102 (I20 probe) or GIP130 (exon I–IV probe) or β-actin mRNAs were used to subsequently probe a premade Northern blot of poly(A+) RNA from the indicated human tissues. Shown are 5-day (I20), 8-day (exons I–IV), and 0.5-day (β-actin) exposures at −70 °C, respectively. The arrowheads and numbers indicate the positions and sizes (kb) of identified mRNA species, whereas the bars and numbers similarly denote RNA markers. D, lysates (50 μg) of HEK 293 cells transfected with either pRK-c-Myc-GIP130 (GIP130), pRK5-c-Myc-FILIP1L-102 (FILIP1L-102), or pRK5-c-Myc vector (control) were analyzed by Western blot with the indicated antibodies. E, schematic alignment of primary structures (bars) of GIP130 and FILIP1L-102 polypeptides expressed in D. Gray shading and black boxes represent coiled-coils and leucine zippers, respectively.
In the presence of [32P-γ]ATP (Fig. 6A), the major 153-kDa polypeptide of rGIP130 was phosphorylated (empty arrowhead), and rGPBP-1 increased 32P incorporation (filled arrowhead), suggesting that rGIP130 stimulates the autophosphorylation of rGPBP-1. This interaction was confirmed through co-immunoprecipitation of the two recombinant proteins from human cells (Fig. 6B). The existence of a mouse ortholog of GIP130 (mGIP130) has been predicted but not reported (GenBankTM accession XR_002318). Based on the XR_002318 sequence, we designed RT-PCR primers and demonstrated the existence of mGIP130 in mice (GenBankTM accession number EU704258). Western blot analysis of muscle extracts and subsequent immunoprecipitation assays demonstrated that mouse striated muscle expressed mGIP130 of 153 kDa and related polypeptides of lower molecular mass (112, 96, and 82 kDa) and that 153-kDa and related 96-kDa polypeptides formed complexes with mGPBP-1, which were precipitated with GPBP-specific antibodies (Fig. 6C). To further analyze the interaction of mGPBP-1 and mGIP130, we performed confocal microscopy studies on isolated myofibrils (Fig. 6D). Whereas mGPBP-1 displayed a more restricted distribution at the M line (see also supplemental Fig. S2), co-localizing with mGIP30 and myosin (arrowheads), mGIP130 co-distributed with myosin within the A band.
FIGURE 6.
GPBP binds and phosphorylates GIP130, a previously unrecognized myofibrillar component. A, 200 ng of yeast FLAG-tagged rGPBP-1 and/or rGIP130 were subjected to phosphorylation and mixtures analyzed by Western blot and autoradiography (32P) during 3 and 16 h and subsequently probed with FLAG-specific antibodies (anti-FLAG). The empty and filled arrowheads denote the positions of rGIP130 and rGPBP-1 primary products, respectively. Numbers and bars at the left indicate size (kDa) and position of individual molecular mass markers. B, HEK 293 cells expressing rGPBP-1 (control) or rGPBP-1- and c-Myc-tagged rGIP130 (GIP130) were lysed and subjected either to Western blot analysis with the indicated antibodies (Lysates) or to immunoprecipitation with anti-c-Myc affinity resin and similarly analyzed (IP). C, protein extracts of mouse diaphragm (50 μg) were analyzed by Western blot (Input) or subjected to immunoprecipitation (500 μg) in the absence (−) or presence of mAb N27 (N27) and similarly analyzed with the indicated antibodies. In B and C, primary and related products are denoted by arrows and their size (kDa). D, myofibrils isolated from mouse diaphragm were analyzed by confocal microscopy for detection of the indicated proteins using rabbit polyclonal antibodies (Myosin), chicken polyclonal antibodies (GPBP-1), and mAb3GIP (GIP). The graphs represent the distribution of fluorescence intensities in the region indicated by the arrow in the merged image. The arrowheads denote co-localization of the three proteins at the M lines of A bands.
GIP130 Promotes Myofibril Myosin Assembly
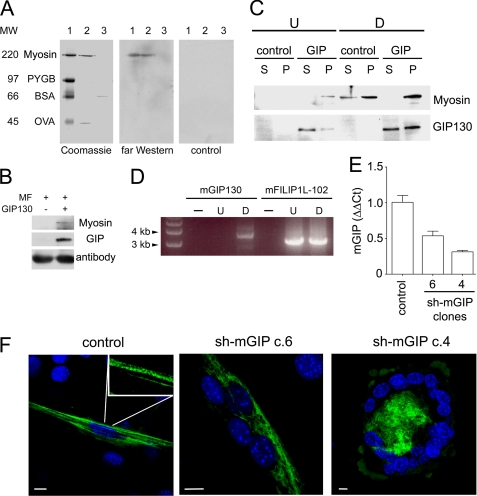
Co-localization of mGIP130 with myosin within myofibrils suggested that GIP130 proteins interact with myosin. This was first explored by far Western procedures using mouse myofibril protein extracts and rGIP130. We observed that rGIP130 bound to mouse skeletal muscle MyoHC present in the extract but also recognized the rabbit skeletal muscle MyoHC, which was included as a molecular mass marker in our electrophoresis standard. The amount of myofibril material used in the study (2 μg) was too low to permit detection of endogenous mGIP130 with mAb3GIP antibodies (Fig. 7A). Subsequently, this interaction was confirmed by pull-down experiments of mouse myofibril extracts using rGIP130 (Fig. 7B). To investigate the biological significance of this interaction in myofibrillogenesis, C2C12 myoblasts expressing rGIP130 were subjected to differentiation and cytoskeleton extraction, and the presence of both myosin and rGIP130 in soluble (S) and insoluble (P) materials was assessed by Western blot (Fig. 7C). In undifferentiated cells, most of rGIP130 material was detected in the soluble fraction; however, and in contrast to control cells, they expressed a limited amount of myosin that was found in the insoluble material along with traces of rGIP130. As expected, differentiation induced myosin expression; however, whereas myosin was found in significant amounts in the soluble fraction in control cells, myosin was found almost exclusively in the insoluble material of rGIP130-expressing myoblasts. Also remarkable was the observation that differentiation was associated with increased presence of rGIP130 in the insoluble fraction, suggesting that rGIP130 bound to soluble myosin and promoted its assembly into myofibrillar bundles. This was further supported by the demonstration that mGIP130 expression was specifically induced early in differentiation (Fig. 7D), and C2C12 cell lines with reduced levels of mGIP130 (Fig. 7E) showed defective myofibril formation (Fig. 7F).
FIGURE 7.
GIP130 binds to myosin and promotes its myofibrillar assembly in differentiating C2C12 cells. A, 2 μg of mouse myofibril protein extract (lanes 2) and 200 ng of BSA (lanes 3) were subjected to SDS-PAGE along with suitable molecular mass markers (lanes 1) and either stained (Coomassie) or transferred to a PVDF membrane and probed with either FLAG-tagged rGIP130 (1 μg/ml) and mAb3GIP antibodies (far Western), or with mAb3GIP antibodies (control). At the left of the composite are the size (kDa) and identities of molecular mass markers: rabbit skeletal muscle MyoHC (Myosin), phosphorylase b (PYGB), bovine serum albumin (BSA), and ovalbumin (OVA). B, myofibril (MF) protein extracts used in A (500 μg) were incubated with (+) or without (−) FLAG-tagged rGIP130 (GIP130), and mixtures were subjected to immunoprecipitation using anti-FLAG antibodies. Precipitates were analyzed by Western blot with suitable antibodies for MyoHC (Myosin) or GIP130 (GIP) detection. IgG heavy chain is shown as a loading control (antibody). C, C2C12 (control) or C2C12 expressing rGIP130 (GIP) cells were subjected to 3-day differentiation. Undifferentiated (U) or differentiated (D) cells were used for cytoskeleton extraction, and supernatants (S) and pellets (P) were analyzed by Western blot using antibodies specific for the indicated proteins. D, the RNA from undifferentiated or 3-day differentiated C2C12 cells was extracted, and the indicated mRNAs were selectively amplified by RT-PCR. Amplification mixtures in the absence (−) or presence of undifferentiated (U) or differentiated (D) RNA were analyzed by agarose gel electrophoresis and stained with ethidium bromide. Indicated are the size and position of DNA markers. E, C2C12 cells were transfected with pSi-mGIP-2 to yield clones differing in mGIP130 expression as assessed by quantitative RT-PCR. Shown are the relative mGIP130 expressions of two sh-mGIP clones (4 and 6) with respect to a clone generated with the corresponding empty vector (control). Shown are the means ± S.D. (error bars). F, the C2C12 clones in E were differentiated for 3 days and analyzed by confocal microscopy for myosin detection. Inset, characteristic striation of myofibrils assembled in differentiated control clone. In contrast, cells of clone 6 displayed loose myofibrils without striation, and cells of clone 4 lacked myofibrils. Bars, 10 μm.
GPBP-1 Regulates the Myofibrillar Assembly of both GIP130 and Myosin
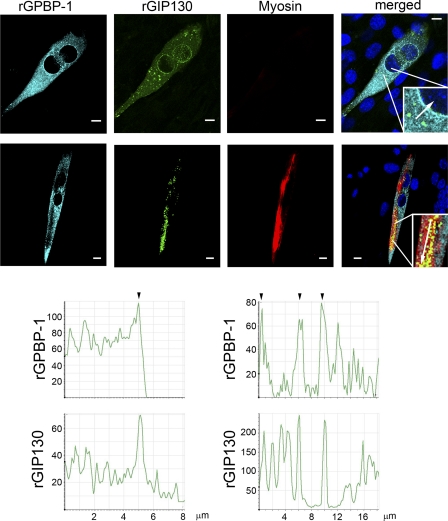
To further investigate the role of GPBP-1 and GIP130 in myogenesis, we expressed their recombinant counterparts in C2C12 cells and subjected these to differentiation and confocal microscopy analysis. Prior to myosin synthesis, rGPBP-1 and rGIP130 formed cytoplasmic aggregates that prominently decorated the nuclear envelope (Fig. 8 top row, left graphs). Expression of myosin promoted recruitment of these aggregates into the nascent myofibrils (Fig. 8, bottom row, right graphs).
FIGURE 8.
GIP130 co-localizes with GPBP-1 and myosin during differentiation. C2C12 myoblasts were co-transfected with individual constructs expressing rGPBP-1 or rGIP130 and then subjected to differentiation. After 1 day (top row) or 3 days (bottom row) of differentiation, cultures were analyzed by confocal microscopy to visualize the indicated proteins. The graphs represent the distribution of fluorescence (cyan and green) intensity in the region indicated by the arrow in the inset of the merged images (top row, left graphs and bottom row, right graphs). The arrowheads denote co-localization of the indicated proteins. Bars, 10 μm.
To determine the role of GPBP-1 in these dynamics, we expressed rGIP130 in gpbp-1+/+ and gpbp-1−/− myoblasts, and cultures were similarly analyzed (Fig. 9A). In gpbp-1+/+ cells, rGIP130 distributed in a linear fashion, co-localizing with skeletal muscle myosin along with developing myofibrils. In contrast, rGIP130 in gpbp-1−/− cells formed large aggregates with myosin that lacked fibrillar organization. These data suggested that GPBP-1 is not required for GIP130 and myosin to associate; however, the organized assembly of these two proteins into myofibrils depends by and large on GPBP-1. This was further confirmed by differentiating gpbp-1−/− myoblasts that expressed rGPBP-1 alone or with rGIP130 (Fig. 9B). Expression of rGPBP-1 promoted fibrillar assembly of myosin in gpbp-1−/− myoblasts and induced migration of rGIP130 toward nascent myofibrillar material. Interestingly, the distribution of myofibrillar material in gpbp-1−/− myoblasts that expressed only rGPBP-1 occupied virtually the central part of cytoplasm, and the cells adopted a preferential spindle-like shape. In contrast, in gpbp-1−/− myoblasts expressing also rGIP130, the myofibrillar material displayed a more peripheral distribution underneath the plasma membrane, and cells displayed a more myotube-like shape, suggesting that cultures expressing both recombinant proteins are more differentiated. Nevertheless, the differences between the two cultures regarding myosin myofibrillar organization were not evident, probably because of the interference of endogenous GIP130 in the cells only expressing rGPBP-1, which is expressed early during myoblast differentiation (see above).
FIGURE 9.
GPBP-1 promotes the myofibrillar assembly of GIP130 and myosin. A, gpbp-1+/+ and gpbp-1−/− myoblasts were transfected with GIP130-EGFP-expressing construct, differentiated for 3 days, and analyzed by confocal microscopy to visualize the indicated proteins. B, gpbp-1−/− myoblasts were transfected with the indicated constructs, and cultures were differentiated for 3 days and analyzed by confocal microscopy for detection of the indicated proteins. In A and B, myosin was detected with MyoHC-specific antibodies, and GIP130-EGFP and EGFP were detected by autofluorescence. In B, GPBP-1 was detected with anti-GPBP-specific antibodies under conditions in which endogenous mGPBP-2 was not detectable Bars, 10 μm.
DISCUSSION
Our previous studies support the notion that targeting of noncollagenous domain-1 by GPBP-1 is pivotal for glomerular basement membrane collagen network organization (2, 5). Here we reveal that the targeting of GIP130 by GPBP-1 is critical for myofibrillar organization in skeletal muscle. GIP130 is a new component of myofibrils, which is related to L-FILIP and FILIP1L-102, previously shown to regulate cell migration and to be down-regulated in ovarian cancer, respectively (26, 27).
Our data also show that accumulation of GPBP-1 in the cytoplasm is accomplished by down-regulating alternative mRNA expression (GPBP-2 and -3). Accordingly, we have not found a net induction of Col4a3bp during C2C12 cell differentiation by quantitative RT-PCR procedures (data not shown). Moreover, these observations provide compelling evidence that alternative mRNA expression (GPBP-2 and -3) is a strategy for extracellular delivery of GPBP-1. The present study also suggests that Col4a3bp expression is critical for myogenesis because knockdown of this gene impairs myoblast differentiation. In contrast, abrogation of GPBP-1 (gpbp-1−/−) does not impair expression of differentiation markers but causes defective myofibril assembly. These observations reveal that GPBP-2 can substitute GPBP-1 deficiency during myoblast differentiation, although it is not as efficient at directing myofibril formation. However, our evidence indicates that under certain circumstances (e.g. development), GPBP-2 can fully replace GPBP-1 and efficiently direct myogenesis and myofibrillar assembly. Thus, the distribution of GPBP-1 and -2 in myofibrils is indistinguishable between gpbp-1+/+ and gpbp-1−/− mice, and gpbp-1−/− mice display no morphological phenotype (supplemental Fig. S2). We have not identified mice expressing GPBP-2 in skeletal muscle (supplemental Fig. S2); however, a human skeletal muscle sample, not associated with a muscular disorder, was previously shown to express exclusively GPBP-2 (6). These observations suggest that there exist in vivo myogenic signals that permit GPBP-2 to compensate functionally when GPBP-1 is not expressed. Similar compensatory mechanisms have been reported in other knock-out models showing normal phenotypes. Thus, ALP (actinin-associated LIM domain protein) was critical for myogenesis in C2C12 cells (28), but mice lacking ALP developed skeletal muscle normally (29). Likewise, the sarcomere-associated protein obscurin was essential for thick filament assembly in cultured myoblasts (30), but mice lacking obscurin evidenced normal sarcomere structure and muscular function (31).
Coordinated down-regulation of GPBP-2 and -3 and accumulation of GPBP-1 in the cytoplasm is an early critical event for myogenic cascade to ensue. Several observations support this notion: 1) mouse myoblast differentiation associates with reduced GPBP-1 secretion (32); 2) knockdown of Col4a3bp expression prevented C2C12 myoblast differentiation (Fig. 2); 3) gpbp-1−/− myoblasts, in contrast to Tg-hGPBP-1 myoblasts, which show accelerated myofibrillogenesis, cannot organize myofibrillar material efficiently (Figs. 3 and 4); 4) gpbp-1−/− myoblasts show defective myotube formation, in contrast to Tg-hGPBP-1 myoblasts, which show enhanced myoblast fusion (Figs. 3 and 4); 5) GIP130, a GPBP-1 downstream effector, binds to myosin and promotes its myofibrillar assembly (Figs. 6 and 7); 6) GIP130 and myosin form disordered aggregates in gpbp-1−/− myoblasts, and this phenotype was reverted, at least in part, by rGPBP-1 expression (Fig. 9); and 7) GPBP-2, which is a less active kinase (6), cannot fully substitute for GPBP-1 during ex vivo mouse myoblast differentiation (Fig. 3). Although transient expression of human GPBP-1 in gpbp-1−/− mouse myoblasts did not fully restore the gpbp-1+/+ phenotype, the partial rescue achieved suggests that GPBP-1 regulates myofibrillogenesis. It is not uncommon to find that defective cell lineages are not fully rescued by the corresponding recombinant protein expression, and several explanations have been proposed (33, 34).
Several lines of evidence have shown that sarcoplasmic reticulum (SR) and sarcomeric structures are linked through protein interactions. Thus, the cytosolic domain of small ankyrin-1, an integral membrane protein of the SR, interacts with the giant protein titin at the level of the Z-disc (35), and also with obscurin at the level of the M line (36). The integral proteins of ER membranes VAP A and B are expressed at the intermyofibrillar SR, displaying a striated distribution. However, within the sarcomere, VAP A mainly associates with A band and VAP B with I band (37). It has been shown that GPBP-2 binds to VAP proteins, displaying a preference for VAP A (38). Phosphorylated GPBP-2 binds to VAP proteins at the ER, and subsequent dephosphorylation promotes its localization at the Golgi apparatus (39). Accordingly, we have found that GPBP-1 and VAP A display a remarkable overlapping distribution at the M line of A band in mouse myocytes, that GPBP-1 and VAP A specifically interact, and that myoblast differentiation induces GPBP-1 (or GPBP-2 in gpbp-1−/− myoblasts) phosphorylation (supplemental Fig. S3). Collectively, the data suggest that in myocytes, a significant amount of GPBP-1 (or GPBP-2 in gpbp-1−/− mice) remains bound to VAP A aligned with the A band.
As for gpbp-1−/− mice, Tg-hGPBP-1 animals do not display any obvious morphological phenotype (supplemental Fig. S4). In contrast, gpbp-1−/− mice did not show a functional phenotype, whereas Tg-hGPBP-1 mice displayed reduced endurance that was not attributable to mitochondrial alterations (supplemental Fig. S5). Interestingly, when Tg-hGPBP-1 mice were allowed to exercise until exhaustion, tubular aggregates were observed in muscle samples (supplemental Fig. S5), suggesting that their low endurance was caused by an alteration in the calcium homeostasis (40). Several lines of evidence suggest that a close and precise spatial relationship between SR and sarcomeric structures is needed for efficient calcium dynamics during contraction-relaxation cycles (41). Accordingly, it has been suggested that obscurin participates in the proper positioning of calcium channels by ensuring adequate alignment of SR and myofibrils through its interaction with small ankyrin 1 (36). Similarly, the association between GPBP-1 and GIP130, by bridging SR and myofibrils, may also participate in positioning calcium channels, and tubular aggregate formation in transgenic mice may result from misalignment due to an excess of free rGPBP-1 displacing GPBP-1-GIP130 from VAP A binding sites. Additionally, GPBP-1 is required for the GIP130-myosin complex to assemble into contractile machinery (Fig. 9), suggesting that an association between VAP A and GPBP-1 is also required for an adequate positioning of the GIP130-myosin complex into the contractile machinery.
Collectively, our observations suggest that myoblast differentiation induces GPBP-1 to accumulate in the cytoplasm and to form aggregates with GIP130. This binary complex binds to SR through VAP A (GPBP-1) and to myosin (GIP130), allowing both myofibrillar formation and SR and sarcomere alignment for efficient calcium dynamics and contractile machinery functionality.
Finally, the evidence presented suggests that GPBP-1 is required not only for myofibril formation but also exerts an additional role in myosin synthesis and myoblast fusion (Figs. 2–4). It has been reported that sphingosine 1-phosphate induces myogenesis (42) and that intracellular production of sphingosine 1-phosphate by sphingosine kinase is critical for myoblast differentiation (43). During differentiation, the intracellular accumulation of GPBP-1 and its phosphorylation (supplemental Fig. S3) most likely reduce interorganellar ceramide transport and increase the local production of sphingosine and its derivative sphingosine 1-phosphate, thereby inducing myogenesis. Moreover, it has been reported that cell fusion and myogenesis also depend on the extracellular matrix components that are secreted by the myoblast (44, 45), including type IV collagen, which is most expressed at the first day of differentiation in C2C12 cells (46). Conceivably, at the early stages of myogenesis, prior to myosin synthesis (Fig. 2), GPBP-1 may critically contribute to basement membrane collagen organization (5) and therefore to adequate cell fusion. Consistently, gpbp-1−/− myoblasts show a deficient cell fusion and myotube formation (Fig. 3). In gpbp-1−/− myoblasts, the increased intracellular expression of GPBP-2 (Fig. 3), a less active kinase (6), cannot fully compensate for the absence of GPBP-1, as reflected by the deficiency in the cytoskeleton organization of myosin (myofibrillogenesis) and actin (myoblast fusion). The latter suggests that supramolecular organization of structural proteins is more dependent on kinase rather than on ceramide transfer activities of GPBP proteins. Consistently, we demonstrate in this report that myofibril formation depends on downstream kinase effector GIP130, and we have also observed that GPBP-1 binds to L-FILIP,4 a GIP130-related protein reported to regulate actin cytoskeleton organization (27).
Supplementary Material
Acknowledgments
We thank the following for technical assistance: Zahara Garzón and Marcos Calderón (general laboratory support), Natalia Palomar (animal care), Alberto Hernández-Cano and Eva María Lafuente (confocal microscopy), and Mario Soriano (electron microscopy).
This work was supported by Ministerio de Ciencia e Innovación (MICINN) Grants SAF2003-09772, SAF2006-12520, and SAF2009-10703 and Generalitat Valenciana Grants GV04B-285, BM-001/2002, ACOMP07-188, and Prometeo 2009/065 (to J. S.); MICINN Grant SAF2008-00011 (to D. B.); Fondo de Investigaciones Sanitarias (FIS) Grants 07/1257 and 07/90043 (to C. N.); and the Programa Torres Quevedo of MICINN (to R. B.) (Spain). This work was also supported in part by National Institutes of Health Grant DK 18381 (to B. G. H.). Juan Saus, Francisco Revert-Ros, and Fernando Revert are partners of Fibrostatin SL, a biotech company exploiting patents related to GPBP.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S5.
F. Revert-Ros, F. Revert, and J. Saus, unpublished observations.
- GPBP
- Goodpasture antigen-binding protein
- ER
- endoplasmic reticulum
- HEK
- human embryonic kidney
- MyoHC
- myosin heavy chain
- SR
- sarcoplasmic reticulum
- mGPBP-1 and mGIP130
- mouse GPBP-1 and GIP130, respectively
- rGPBP-1
- rGIP130, and rFILIP1L-102, recombinant GPBP-1, GIP130 and FILIP1L-102, respectively.
REFERENCES
- 1. Revert F., Ventura I., Martínez-Martínez P., Granero-Moltó F., Revert-Ros F., Macías J., Saus J. (2008) J. Biol. Chem. 283, 30246–30255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raya A., Revert F., Navarro S., Saus J. (1999) J. Biol. Chem. 274, 12642–12649 [DOI] [PubMed] [Google Scholar]
- 3. Saus J., Wieslander J., Langeveld J. P., Quinones S., Hudson B. G. (1988) J. Biol. Chem. 263, 13374–13380 [PubMed] [Google Scholar]
- 4. Hudson B. G., Tryggvason K., Sundaramoorthy M., Neilson E. G. (2003) N. Engl. J. Med. 348, 2543–2556 [DOI] [PubMed] [Google Scholar]
- 5. Revert F., Merino R., Monteagudo C., Macias J., Peydró A., Alcácer J., Muniesa P., Marquina R., Blanco M., Iglesias M., Revert-Ros F., Merino J., Saus J. (2007) Am. J. Pathol. 171, 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raya A., Revert-Ros F., Martinez-Martinez P., Navarro S., Rosello E., Vieites B., Granero F., Forteza J., Saus J. (2000) J. Biol. Chem. 275, 40392–40399 [DOI] [PubMed] [Google Scholar]
- 7. Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. (2003) Nature 426, 803–809 [DOI] [PubMed] [Google Scholar]
- 8. Fugmann T., Hausser A., Schöffler P., Schmid S., Pfizenmaier K., Olayioye M. A. (2007) J. Cell Biol. 178, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rao R. P., Yuan C., Allegood J. C., Rawat S. S., Edwards M. B., Wang X., Merrill A. H., Jr., Acharya U., Acharya J. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Granero-Moltó F., Sarmah S., O'Rear L., Spagnoli A., Abrahamson D., Saus J., Hudson B. G., Knapik E. W. (2008) J. Biol. Chem. 283, 20495–20504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang X., Rao R. P., Kosakowska-Cholody T., Masood M. A., Southon E., Zhang H., Berthet C., Nagashim K., Veenstra T. K., Tessarollo L., Acharya U., Acharya J. K. (2009) J. Cell Biol. 184, 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Penadés J. R., Bernal D., Revert F., Johansson C., Fresquet V. J., Cervera J., Wieslander J., Quinones S., Saus J. (1995) Eur. J. Biochem. 229, 754–760 [DOI] [PubMed] [Google Scholar]
- 13. Miralem T., Gibbs P. E., Revert F., Saus J., Maines M. D. (2010) J. Biol. Chem. 285, 12551–12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frock R. L., Kudlow B. A., Evans A. M., Jameson S. A., Hauschka S. D., Kennedy B. K. (2006) Genes Dev. 20, 486–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGrath M. J., Cottle D. L., Nguyen M. A., Dyson J. M., Coghill I. D., Robinson P. A., Holdsworth M., Cowling B. S., Hardeman E. C., Mitchell C. A., Brown S. (2006) J. Biol. Chem. 281, 7666–7683 [DOI] [PubMed] [Google Scholar]
- 16. Clark K., Middelbeek J., Lasonder E., Dulyaninova N. G., Morrice N. A., Ryazanov A. G., Bresnick A. R., Figdor C. G., van Leeuwen F. N. (2008) J. Mol. Biol. 378, 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yaffe D., Saxel O. (1977) Nature 270, 725–727 [DOI] [PubMed] [Google Scholar]
- 18. Davis R. L., Weintraub H., Lassar A. B. (1987) Cell. 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- 19. Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., Nabeshima Y. (1993) Nature 364, 532–535 [DOI] [PubMed] [Google Scholar]
- 20. Diel P., Baadners D., Schlüpmann K., Velders M., Schwarz J. P. (2008) J. Mol. Endocrinol. 40, 231–241 [DOI] [PubMed] [Google Scholar]
- 21. Calera M. R., Pilch P. F. (1998) Biochem. Biophys. Res. Commun. 251, 835–841 [DOI] [PubMed] [Google Scholar]
- 22. Vandromme M., Rochat A., Meier R., Carnac G., Besser D., Hemmings B. A., Fernandez A., Lamb N. J. (2001) J. Biol. Chem. 276, 8173–8179 [DOI] [PubMed] [Google Scholar]
- 23. Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 24. Galbiati F., Volonte D., Engelman J. A., Scherer P. E., Lisanti M. P. (1999) J. Biol. Chem. 274, 30315–30321 [DOI] [PubMed] [Google Scholar]
- 25. Zeschnigk M., Kozian D., Kuch C., Schmoll M., Starzinski-Powitz A. (1995) J. Cell Sci. 108, 2973–2981 [DOI] [PubMed] [Google Scholar]
- 26. Nagano T., Yoneda T., Hatanaka Y., Kubota C., Murakami F., Sato M. (2002) Nat. Cell Biol. 4, 495–501 [DOI] [PubMed] [Google Scholar]
- 27. Mok S. C., Wong K. K., Chan R. K., Lau C. C., Tsao S. W., Knapp R. C., Berkowitz R. S. (1994) Gynecol. Oncol. 52, 247–252 [DOI] [PubMed] [Google Scholar]
- 28. Pomiès P., Pashmforoush M., Vegezzi C., Chien K. R., Auffray C., Beckerle M. C. (2007) Mol. Biol. Cell 18, 1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jo K., Rutten B., Bunn R. C., Bredt D. S. (2001) Mol. Cell. Biol. 21, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ackermann M. A., Hu L. Y., Bowman A. L., Bloch R. J., Kontrogianni-Konstantopoulos A. (2009) Mol. Biol. Cell 20, 2963–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lange S., Ouyang K., Meyer G., Cui L., Cheng H., Lieber R. L., Chen J. (2009) J. Cell Sci. 122, 2640–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Henningsen J., Rigbolt K. T., Blagoev B., Pedersen B. K., Kratchmarova I. (2010) Mol. Cell. Proteomics 9, 2482–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Broers J. L., Peeters E. A., Kuijpers H. J., Endert J., Bouten C. V., Oomens C. W., Baaijens F. P., Ramaekers F. C. (2004) Hum. Mol Genet. 13, 2567–2580 [DOI] [PubMed] [Google Scholar]
- 34. Rotwein P., Wilson E. M. (2009) J. Cell. Physiol. 219, 503–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kontrogianni-Konstantopoulos A., Bloch R. J. (2003) J. Biol. Chem. 278, 3985–3991 [DOI] [PubMed] [Google Scholar]
- 36. Bagnato P., Barone V., Giacomello E., Rossi D., Sorrentino V. (2003) J. Cell Biol. 160, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gkogkas C., Middleton S., Kremer A. M., Wardrope C., Hannah M., Gillingwater T. H., Skehel P. (2008) Hum. Mol. Genet. 17, 1517–1526 [DOI] [PubMed] [Google Scholar]
- 38. Kawano M., Kumagai K., Nishijima M., Hanada K. (2006) J. Biol. Chem. 281, 30279–30288 [DOI] [PubMed] [Google Scholar]
- 39. Saito S., Matsui H., Kawano M., Kumagai K., Tomishige N., Hanada K., Echigo S., Tamura S., Kobayashi T. (2008) J. Biol. Chem. 283, 6584–6593 [DOI] [PubMed] [Google Scholar]
- 40. Salviati G., Pierobon-Bormioli S., Betto R., Damiani E., Angelini C., Ringel S. P., Salvatori S., Margreth A. (1985) Muscle Nerve 8, 299–306 [DOI] [PubMed] [Google Scholar]
- 41. Takekura H., Flucher B. E., Franzini-Armstrong C. (2001) Dev. Biol. 239, 204–214 [DOI] [PubMed] [Google Scholar]
- 42. Donati C., Meacci E., Nuti F., Becciolini L., Farnararo M., Bruni P. (2005) FASEB J. 19, 449–451 [DOI] [PubMed] [Google Scholar]
- 43. Meacci E., Nuti F., Donati C., Cencetti F., Farnararo M., Bruni P. (2008) J. Cell. Physiol. 214, 210–220 [DOI] [PubMed] [Google Scholar]
- 44. Nandan D., Clarke E. P., Ball E. H., Sanwal B. D. (1990) J. Cell Biol. 110, 1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Osses N., Brandan E. (2002) Am. J. Physiol. Cell Physiol. 282, C383–C394 [DOI] [PubMed] [Google Scholar]
- 46. Moran J. L., Li Y., Hill A. A., Mounts W. M., Miller C. P. (2002) Physiol. Genomics 10, 103–111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.