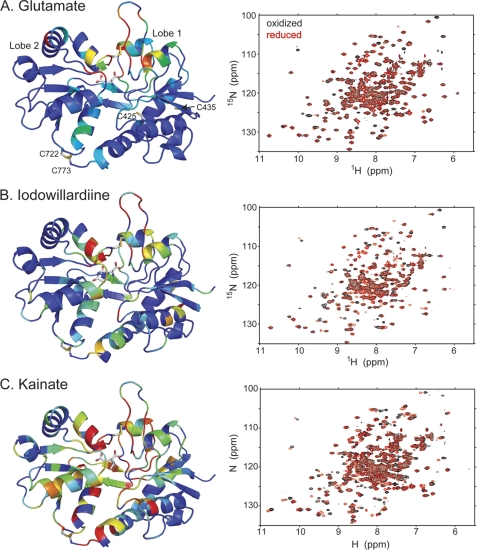
FIGURE 1.
Comparison of the amide backbone chemical shifts in the oxidized and reduced (DTT) form of A452C/S652C mutant of GluA2 LBD bound to glutamate (A; Protein Data Bank code 3T93), iodowillardiine (B; Protein Data Bank code 3T96), and kainate (C; Protein Data Bank code 3T9H). On the right is shown the 1H,15N-HSQC spectrum, and on the left, a representation of the resonances that have shifted upon reduction of the A452C/S652C disulfide bond is shown. The oxidized form is identical with no additions and in the presence of Cu-phenanthroline, suggesting that the disulfide bond forms spontaneously. The difference in chemical shift between the oxidized and reduced from was quantitated using the formula, chemical shift difference = . Residues were assigned colors with a chemical shift difference between 0 and 5 assigned to a rainbow of colors from blue to red.