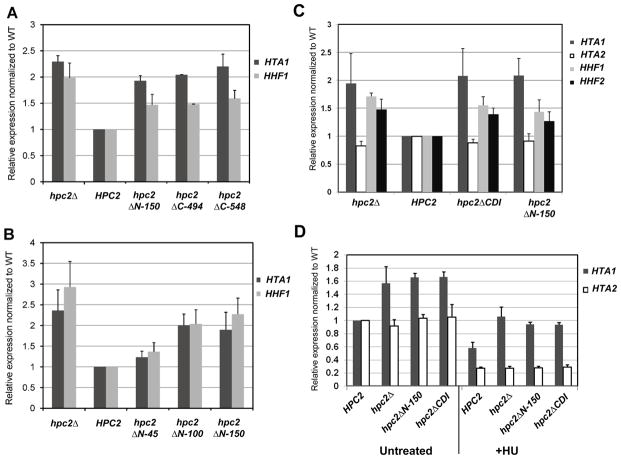
Figure 3. The Hpc2 N-terminal conserved domain is essential for histone gene repression.
(A) N-terminal- and C-terminal Hpc2 truncations cause derepression of the histone genes similarly to hpc2Δ. YPP996 strain was transformed with the indicated YCp:URA3 plasmid and grown in SD Ura- media, total RNA was extracted and HTA1 and HHF1 mRNA expression were tested by RT-qPCR. (B) The Hpc2 region between amino acid 100 and 150 is responsible for the depression of the histone genes. The strain and method are the same as in (A). (C) The Hpc2 CDI conserved domain is responsible for the derepression of the histone genes observed in an Hpc2 N-terminal truncated mutant. YPP996 strain was transformed with the indicated YIp:TRP1 plasmid and, after selection, were grown in YPD. RNA extraction and RT-qPCR were carried out as in (A), to check the level of HTA1, HTA2, HHF1, and HHF2. (D) Hpc2 CDI domain is necessary for repression of histone genes upon HU (hydroxyurea) treatment. The same strains were used as in (C). Cell were grown to log phase and collected from untreated culture or subjected to HU treatment (200mM) for 45min. RT-qPCR was used to monitor the level of HTA1 and HTA2 expression. HTA2 is used as a HIR-independent histone gene control. All the RT-qPCR were normalized to ACT1 expression level.