Abstract
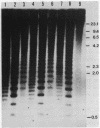
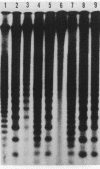
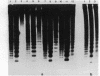
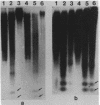
The distribution of 5-methyl cytosine (5-MeC) residues in a highly repetitive sequence, mouse major satellite, was examined in germinal versus somatic DNAs by digestion with the methylation sensitive isoschizomers Msp I and Hpa II and Southern blot analysis, using a cloned satellite probe. DNA from liver, brain, and a mouse fibroblast cell line, C3H 10T1/2, yielded a multimeric hybridization pattern after digestion with Msp I (and control Eco RI) but were resistant to digestion with Hpa II, reflecting a high level of methylation of the satellite sequences. In contrast, DNA from mature sperm was undermethylated at these same sequences as indicated by the ability of Hpa II to generate a multimeric pattern. DNAs from purified populations of testis cells in different stages of spermatogenesis were examined to determine when during germ cell differentiation the undermethylation was established. As early as in primitive type A, type A, and type B spermatogonia, an undermethylation of satellite sequences was observed. This suggest that this highly specific undermethylation of germ cell satellite DNA occurs very early in the germ cell lineage, prior to entry into meiosis.
Full text
PDF




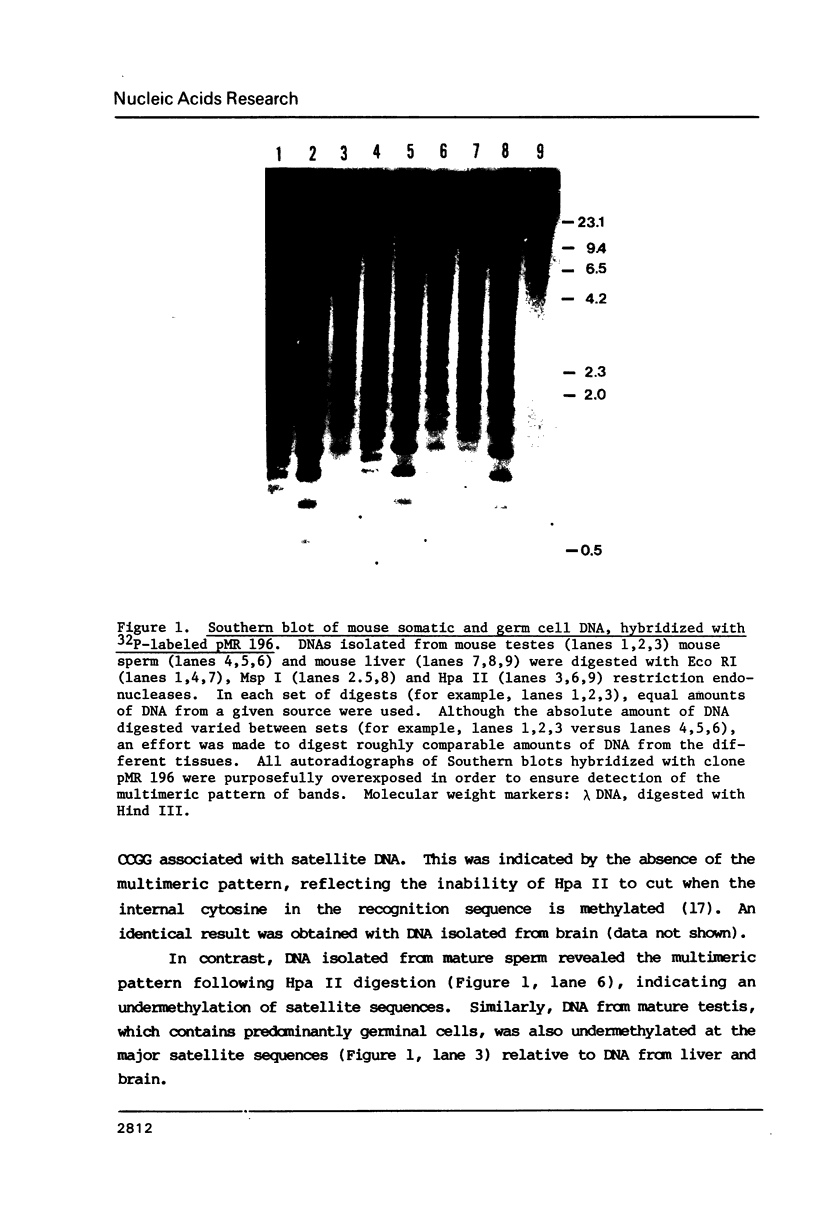

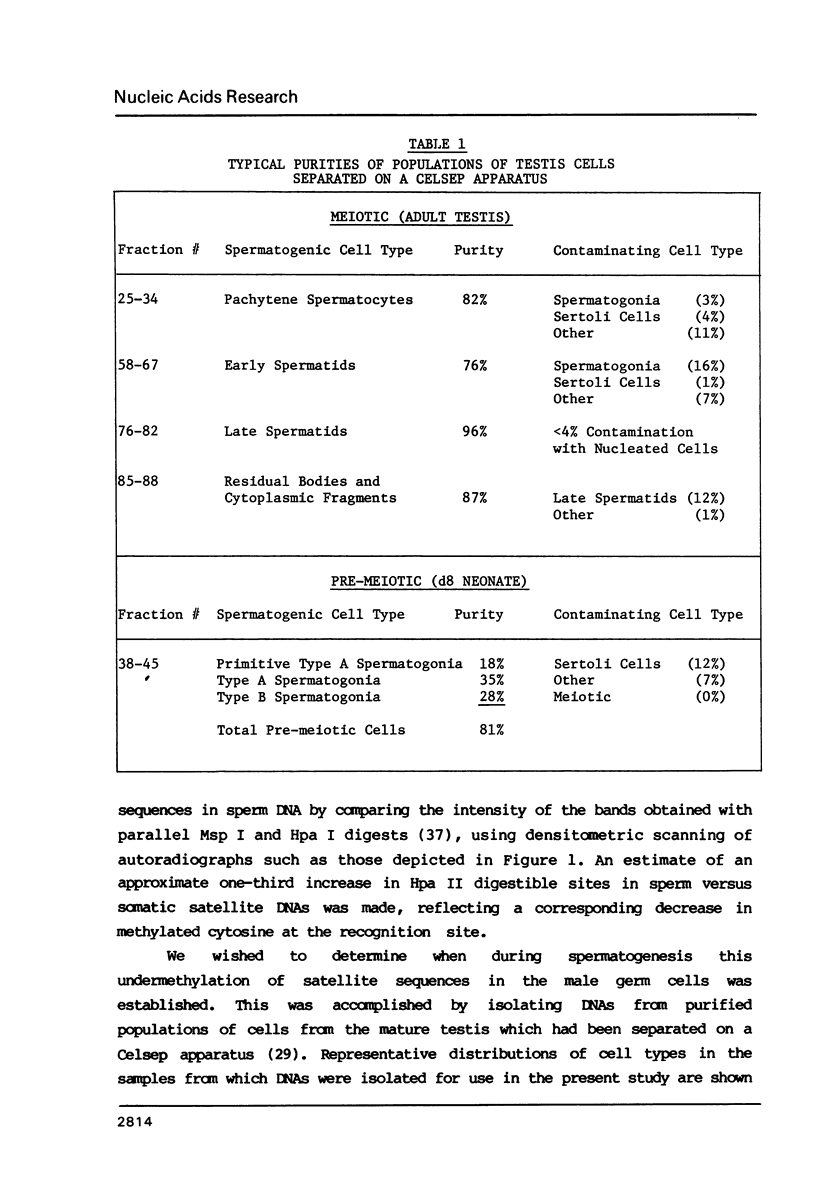








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., Burdon R. H., Fulton J. Methylation of satellite DNA. Biochem Biophys Res Commun. 1983 Jun 15;113(2):695–702. doi: 10.1016/0006-291x(83)91782-5. [DOI] [PubMed] [Google Scholar]
- Bellvé A. R., Cavicchia J. C., Millette C. F., O'Brien D. A., Bhatnagar Y. M., Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977 Jul;74(1):68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé A. R., Millette C. F., Bhatnagar Y. M., O'Brien D. A. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem. 1977 Jul;25(7):480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A., Taggart M., Macleod D. Loss of rDNA methylation accompanies the onset of ribosomal gene activity in early development of X. laevis. Cell. 1981 Nov;26(3 Pt 1):381–390. doi: 10.1016/0092-8674(81)90207-5. [DOI] [PubMed] [Google Scholar]
- Cedar H., Solage A., Glaser G., Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6(6):2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman V., Forrester L., Sanford J., Hastie N., Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984 Jan 19;307(5948):284–286. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- Chen C. L., Dionne F. T., Roberts J. L. Regulation of the pro-opiomelanocortin mRNA levels in rat pituitary by dopaminergic compounds. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2211–2215. doi: 10.1073/pnas.80.8.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Lopata M. A., MacDonald R. J., Cowan N. J., Rutter W. J., Kirschner M. W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980 May;20(1):95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- Cohen A. K., Huh T. Y., Helleiner C. W. Transcription of satellite DNA in mouse L-cells. Can J Biochem. 1973 May;51(5):529–532. doi: 10.1139/o73-065. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Posakony J. W. Repetitive sequence transcripts in development. Nature. 1982 Jun 24;297(5868):633–635. doi: 10.1038/297633a0. [DOI] [PubMed] [Google Scholar]
- Diaz M. O., Barsacchi-Pilone G., Mahon K. A., Gall J. G. Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell. 1981 Jun;24(3):649–659. doi: 10.1016/0092-8674(81)90091-x. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation--a regulatory signal in eukaryotic gene expression. J Gen Virol. 1981 Nov;57(Pt 1):1–20. doi: 10.1099/0022-1317-57-1-1. [DOI] [PubMed] [Google Scholar]
- Ellis T. H., Goldsbrough P. B. The analysis of the distribution of restriction endonuclease sites in repetitive DNAs. Nucleic Acids Res. 1981 Apr 10;9(7):1551–1558. doi: 10.1093/nar/9.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Size and structure of the highly repetitive BAM HI element in mice. Nucleic Acids Res. 1983 Aug 11;11(15):5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983 Jun 24;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Gebhard W., Meitinger T., Höchtl J., Zachau H. G. A new family of interspersed repetitive DNA sequences in the mouse genome. J Mol Biol. 1982 May 25;157(3):453–471. doi: 10.1016/0022-2836(82)90471-5. [DOI] [PubMed] [Google Scholar]
- Harbers K., Harbers B., Spencer J. H. Nucleotide clusters in deoxyribonucleic acids. XII. The distribution of 5-methylcytosine in pyrimidine oligonucleotides of mouse L-cell satellite DNA and main band DNA. Biochem Biophys Res Commun. 1975 Sep 16;66(2):738–746. doi: 10.1016/0006-291x(75)90572-0. [DOI] [PubMed] [Google Scholar]
- Harel J., Hanania N., Tapiero H., Harel L. RNA replication by nuclear satellite DNA in different mouse cells. Biochem Biophys Res Commun. 1968 Nov 25;33(4):696–701. doi: 10.1016/0006-291x(68)90352-5. [DOI] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981 Feb 11;9(3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. W. Chromosomal and nuclear location of mouse satellite DNA in individual cells. Nature. 1970 Mar 7;225(5236):912–915. doi: 10.1038/225912a0. [DOI] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Keshet E., Cedar H. Effect of CpG methylation on Msp I. Nucleic Acids Res. 1983 Jun 11;11(11):3571–3580. doi: 10.1093/nar/11.11.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders K. K., Paterson B. M. A short interspersed repetitive element found near some mouse structural genes. Nucleic Acids Res. 1982 Dec 11;10(23):7715–7729. doi: 10.1093/nar/10.23.7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon C., Ohkubo H., Pastan I., de Crombrugghe B. Unusual methylation pattern of the alpha 2 (l) collagen gene. Cell. 1982 May;29(1):203–210. doi: 10.1016/0092-8674(82)90104-0. [DOI] [PubMed] [Google Scholar]
- Miller O. J., Schnedl W., Allen J., Erlanger B. F. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974 Oct 18;251(5476):636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- O'Brien D. A., Bellvé A. R. Protein constituents of the mouse spermatozoon. I. An electrophoretic characterization. Dev Biol. 1980 Mar 15;75(2):386–404. doi: 10.1016/0012-1606(80)90171-2. [DOI] [PubMed] [Google Scholar]
- OAKBERG E. F. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat. 1956 Nov;99(3):391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Gall J. G. Chromosomal localization of mouse satellite DNA. Science. 1970 Jun 12;168(3937):1356–1358. doi: 10.1126/science.168.3937.1356. [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Lohe A. R., Gerlach W. L., Dunsmuir P., Dennis E. S., Appels R. Fine structure and evolution of DNA in heterochromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1121–1135. doi: 10.1101/sqb.1978.042.01.113. [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Miklos G. L., Goodchild D. J. Sex chromosome meiotic drive systems in Drosophila melanogaster I. Abnormal spermatid development in males with a heterochromatin-deficient X chromosome (sc-4sc-8). Genetics. 1975 Apr;79(4):613–634. doi: 10.1093/genetics/79.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Reilly J. G., Thomas C. A., Jr, Sen A. DNA methylation in mouse cells in culture as measured by restriction enzymes. Biochim Biophys Acta. 1982 Apr 26;697(1):53–59. doi: 10.1016/0167-4781(82)90044-6. [DOI] [PubMed] [Google Scholar]
- Romrell L. J., Bellvé A. R., Fawcett D. W. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol. 1976 Mar;49(1):119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- Russell G. J., Walker P. M., Elton R. A., Subak-Sharpe J. H. Doublet frequency analysis of fractionated vertebrate nuclear DNA. J Mol Biol. 1976 Nov;108(1):1–23. doi: 10.1016/s0022-2836(76)80090-3. [DOI] [PubMed] [Google Scholar]
- Salomon R., Kaye A. M., Herzberg M. Mouse nuclear satellite DNA: 5-methylcytosine content, pyrimidine isoplith distribution and electron microscopic appearance. J Mol Biol. 1969 Aug 14;43(3):581–592. doi: 10.1016/0022-2836(69)90360-x. [DOI] [PubMed] [Google Scholar]
- Sano H., Sager R. Tissue specificity and clustering of methylated cystosines in bovine satellite I DNA. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3584–3588. doi: 10.1073/pnas.79.11.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi D. V., Garrett C. E., Barr P. J. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983 May;33(1):9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Riggs A. D. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979 Mar 9;203(4384):1019–1021. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- Siracusa L. D., Chapman V. M., Bennett K. L., Hastie N. D., Pietras D. F., Rossant J. Use of repetitive DNA sequences to distinguish Mus musculus and Mus caroli cells by in situ hybridization. J Embryol Exp Morphol. 1983 Feb;73:163–178. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sturm K. S., Taylor J. H. Distribution of 5-methylcytosine in the DNA of somatic and germline cells from bovine tissues. Nucleic Acids Res. 1981 Sep 25;9(18):4537–4546. doi: 10.1093/nar/9.18.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley J. M., Macgregor H. C., Erba H. P. Satellite DNA is transcribed on lampbrush chromosomes. Nature. 1980 Feb 14;283(5748):686–688. doi: 10.1038/283686a0. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock R., Sweet R., Weiss M., Cedar H., Axel R. Intragenic DNA spacers interrupt the ovalbumin gene. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1299–1303. doi: 10.1073/pnas.75.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]