Abstract
The major and the minor satellite sequences of Mus musculus were undermethylated in both sperm and oocyte DNAs relative to the amount of undermethylation observed in adult somatic tissue DNA. This hypomethylation was specific for satellite sequences in sperm DNA. Dispersed repetitive and low copy sequences show a high degree of methylation in sperm DNA; however, a dispersed repetitive sequence was undermethylated in oocyte DNA. This finding suggests a difference in the amount of total genomic DNA methylation between sperm and oocyte DNA. The methylation levels of the minor satellite sequences did not change during spermiogenesis, and were not associated with the onset of meiosis or a specific stage in sperm development.
Full text
PDF




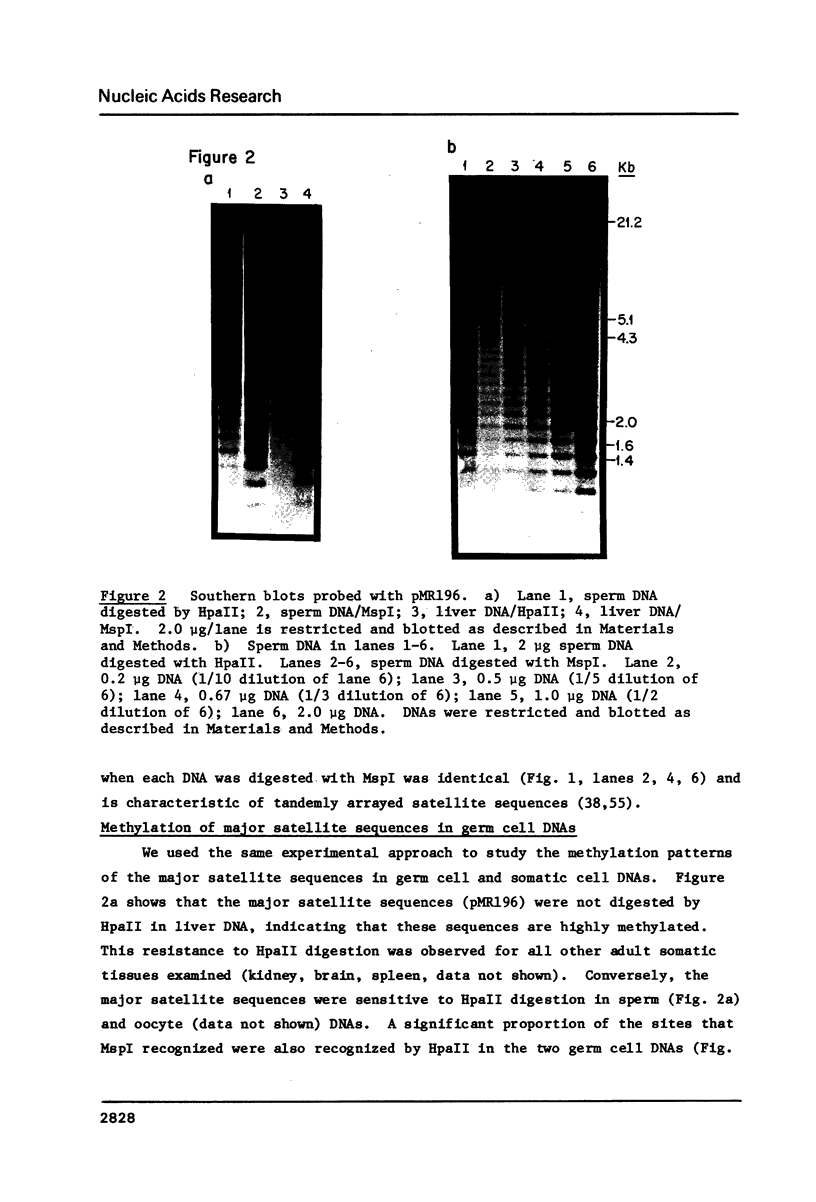








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Brown S. D., Dover G. Organization and evolutionary progress of a dispersed repetitive family of sequences in widely separated rodent genomes. J Mol Biol. 1981 Aug 25;150(4):441–466. doi: 10.1016/0022-2836(81)90374-0. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Wagar M. A., Atkinson M. R. Calcium-dependent priming of DNA synthesis in isolated rat liver nuclei. Biochem Biophys Res Commun. 1970 Apr 24;39(2):254–259. doi: 10.1016/0006-291x(70)90786-2. [DOI] [PubMed] [Google Scholar]
- Busslinger M., deBoer E., Wright S., Grosveld F. G., Flavell R. A. The sequence GGCmCGG is resistant to MspI cleavage. Nucleic Acids Res. 1983 Jun 11;11(11):3559–3569. doi: 10.1093/nar/11.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Solage A., Glaser G., Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6(6):2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandley A. C., Hotta Y., Stern H. Biochemical analysis of meiosis in the male mouse. I. Separation of DNA labelling of specific spermatogenic stages. Chromosoma. 1977 Jul 8;62(3):243–253. doi: 10.1007/BF00286046. [DOI] [PubMed] [Google Scholar]
- Chapman V. M., Kratzer P. G., Siracusa L. D., Quarantillo B. A., Evans R., Liskay R. M. Evidence for DNA modification in the maintenance of X-chromosome inactivation of adult mouse tissues. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5357–5361. doi: 10.1073/pnas.79.17.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman V., Forrester L., Sanford J., Hastie N., Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984 Jan 19;307(5948):284–286. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- Cheng S. M., Schildkraut C. L. A family of moderately repetitive sequences in mouse DNA. Nucleic Acids Res. 1980 Sep 25;8(18):4075–4090. doi: 10.1093/nar/8.18.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C. Methylation of milk-borne and genetically transmitted mouse mammary tumor virus proviral DNA. Cell. 1980 Mar;19(3):653–662. doi: 10.1016/s0092-8674(80)80042-0. [DOI] [PubMed] [Google Scholar]
- Cooper D. N. Eukaryotic DNA methylation. Hum Genet. 1983;64(4):315–333. doi: 10.1007/BF00292363. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Mulder C., Fleckenstein B. Methylation of Herpesvirus saimiri DNA in lymphoid tumor cell lines. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3839–3843. doi: 10.1073/pnas.76.8.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Donahue R. P. Cytogenetic analysis of the first cleavage division in mouse embryos (fertilization-pronuclei-T163H translocation). Proc Natl Acad Sci U S A. 1972 Jan;69(1):74–77. doi: 10.1073/pnas.69.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Size and structure of the highly repetitive BAM HI element in mice. Nucleic Acids Res. 1983 Aug 11;11(15):5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard C., Doly J., Cortadas J., Bernardi G. The primary structure of bovine satellite 1.715. Nucleic Acids Res. 1981 Nov 25;9(22):6069–6082. doi: 10.1093/nar/9.22.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983 Jun 24;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Wang R. Y., Kuo K. C., Gehrke C. W., Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983 May 25;11(10):3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Groudine M., Weintraub H. Activation of globin genes during chicken development. Cell. 1981 May;24(2):393–401. doi: 10.1016/0092-8674(81)90329-9. [DOI] [PubMed] [Google Scholar]
- Guntaka R. V., Rao P. Y., Mitsialis S. A., Katz R. Modification of avian sarcoma proviral DNA sequences in nonpermissive XC cells but not in permissive chicken cells. J Virol. 1980 May;34(2):569–572. doi: 10.1128/jvi.34.2.569-572.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper M. I., Fosten M., Monk M. Preferential paternal X inactivation in extraembryonic tissues of early mouse embryos. J Embryol Exp Morphol. 1982 Feb;67:127–135. [PubMed] [Google Scholar]
- Heller R., Arnheim N. Structure and organization of the highly repeated and interspersed 1.3 kb EcoRI-Bg1II sequence family in mice. Nucleic Acids Res. 1980 Nov 11;8(21):5031–5042. doi: 10.1093/nar/8.21.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981 Feb 11;9(3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Zachau H. G. Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur J Biochem. 1977 Mar 1;73(2):383–392. doi: 10.1111/j.1432-1033.1977.tb11329.x. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaput J., Sneider T. W. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions. Nucleic Acids Res. 1979 Dec 20;7(8):2303–2322. doi: 10.1093/nar/7.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet E., Cedar H. Effect of CpG methylation on Msp I. Nucleic Acids Res. 1983 Jun 11;11(11):3571–3580. doi: 10.1093/nar/11.11.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D. Fluorescence spot tests for DNA endonuclease, ligase, and topoisomerase activities. Anal Biochem. 1980 Sep 15;107(2):311–313. doi: 10.1016/0003-2697(80)90388-7. [DOI] [PubMed] [Google Scholar]
- Kratzer P. G., Chapman V. M., Lambert H., Evans R. E., Liskay R. M. Differences in the DNA of the inactive X chromosomes of fetal and extraembryonic tissues of mice. Cell. 1983 May;33(1):37–42. doi: 10.1016/0092-8674(83)90332-x. [DOI] [PubMed] [Google Scholar]
- Lubit B. W., Pham T. D., Miller O. J., Erlanger B. F. Localization of 5-methylcytosine in human metaphase chromosomes by immunoelectron microscopy. Cell. 1976 Dec;9(4 Pt 1):503–509. doi: 10.1016/0092-8674(76)90032-5. [DOI] [PubMed] [Google Scholar]
- Lubit B. W., Schreck R. R., Miller O. J., Erlanger B. F. Human chromosome structure as revealed by an immunoperoxidase staining procedure. Exp Cell Res. 1974 Dec;89(2):426–429. doi: 10.1016/0014-4827(74)90815-5. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Miller O. J., Schnedl W., Allen J., Erlanger B. F. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974 Oct 18;251(5476):636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- Naveh-Many T., Cedar H. Active gene sequences are undermethylated. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4246–4250. doi: 10.1073/pnas.78.7.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer A., Wigler M., Axel R., Silverstein S. The transfer and stable integration of the HSV thymidine kinase gene into mouse cells. Cell. 1978 May;14(1):133–141. doi: 10.1016/0092-8674(78)90308-2. [DOI] [PubMed] [Google Scholar]
- Pietras D. F., Bennett K. L., Siracusa L. D., Woodworth-Gutai M., Chapman V. M., Gross K. W., Kane-Haas C., Hastie N. D. Construction of a small Mus musculus repetitive DNA library: identification of a new satellite sequence in Mus musculus. Nucleic Acids Res. 1983 Oct 25;11(20):6965–6983. doi: 10.1093/nar/11.20.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahe B., Erickson R. P., Quinto M. Methylation of unique sequence DNA during spermatogenesis in mice. Nucleic Acids Res. 1983 Nov 25;11(22):7947–7959. doi: 10.1093/nar/11.22.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H. Distribution of 5-methylcytosine in chromatin. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2725–2728. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon R., Kaye A. M., Herzberg M. Mouse nuclear satellite DNA: 5-methylcytosine content, pyrimidine isoplith distribution and electron microscopic appearance. J Mol Biol. 1969 Aug 14;43(3):581–592. doi: 10.1016/0022-2836(69)90360-x. [DOI] [PubMed] [Google Scholar]
- Schnedl W., Dev V. G., Tantravahi R., Miller D. A., Erlanger B. F., Miller O. J. 5-Methylcytosine in heterochromatic regions of chromosomes: chimpanzee and gorilla compared to the human. Chromosoma. 1975 Sep 15;52(1):59–66. doi: 10.1007/BF00285789. [DOI] [PubMed] [Google Scholar]
- Shen C. K., Maniatis T. Tissue-specific DNA methylation in a cluster of rabbit beta-like globin genes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6634–6638. doi: 10.1073/pnas.77.11.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Riggs A. D. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979 Mar 9;203(4384):1019–1021. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- Solage A., Cedar H. Organization of 5-methylcytosine in chromosomal DNA. Biochemistry. 1978 Jul 11;17(14):2934–2938. doi: 10.1021/bi00607a036. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Sturm K. S., Taylor J. H. Distribution of 5-methylcytosine in the DNA of somatic and germline cells from bovine tissues. Nucleic Acids Res. 1981 Sep 25;9(18):4537–4546. doi: 10.1093/nar/9.18.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc Natl Acad Sci U S A. 1980 Jan;77(1):253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N., Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975 Aug 21;256(5519):640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Neumann R., Kuhlmann I., Sutter D., Doerfler W. DNA methylation and viral gene expression in adenovirus-transformed and -infected cells. Nucleic Acids Res. 1980 Jun 11;8(11):2461–2473. doi: 10.1093/nar/8.11.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliva C. F., Jahn C. L., Comer M. B., Hutchison C. A., 3rd, Edgell M. H. The L1Md long interspersed repeat family in the mouse: almost all examples are truncated at one end. Nucleic Acids Res. 1983 Dec 20;11(24):8847–8859. doi: 10.1093/nar/11.24.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Larsen A., Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981 May;24(2):333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- West J. D., Frels W. I., Chapman V. M., Papaioannou V. E. Preferential expression of the maternally derived X chromosome in the mouse yolk sac. Cell. 1977 Dec;12(4):873–882. doi: 10.1016/0092-8674(77)90151-9. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Groffen J., Flavell R. A. A novel type of secondary modification of two CCGG residues in the human gamma delta beta-globin gene locus. Nucleic Acids Res. 1980 Oct 24;8(20):4563–4574. doi: 10.1093/nar/8.20.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]