Abstract
Nitrite-dependent anaerobic oxidation of methane (n-damo) and ammonium (anammox) are two recently discovered processes in the nitrogen cycle that are catalyzed by n-damo bacteria, including “Candidatus Methylomirabilis oxyfera,” and anammox bacteria, respectively. The feasibility of coculturing anammox and n-damo bacteria is important for implementation in wastewater treatment systems that contain substantial amounts of both methane and ammonium. Here we tested this possible coexistence experimentally. To obtain such a coculture, ammonium was fed to a stable enrichment culture of n-damo bacteria that still contained some residual anammox bacteria. The ammonium supplied to the reactor was consumed rapidly and could be gradually increased from 1 to 20 mM/day. The enriched coculture was monitored by fluorescence in situ hybridization and 16S rRNA and pmoA gene clone libraries and activity measurements. After 161 days, a coculture with about equal amounts of n-damo and anammox bacteria was established that converted nitrite at a rate of 0.1 kg-N/m3/day (17.2 mmol day−1). This indicated that the application of such a coculture for nitrogen removal may be feasible in the near future.
INTRODUCTION
Under anaerobic conditions, nitrite serves as the terminal electron acceptor for microorganisms that oxidize ammonium and methane. The anaerobic ammonium oxidation (anammox) process has already been recognized as a cost-effective and environment-friendly alternative to conventional nitrogen removal systems (14). Different configurations of anammox reactors are successfully applied in full-scale sewage treatment plants to treat highly ammonium-loaded wastewater such as anaerobic digester effluents (1, 30).
Ammonium and methane are major end products of anaerobic digestion, where the latter is recycled and used as an energy source, generally for electricity production. Methane in the gas phase can be collected and purified easily, but dissolved methane is difficult to recover, especially at lower temperatures. The slow release of the dissolved methane into the environment contributes to the greenhouse effect (4, 5) and is therefore undesirable. Methane is an energy-rich compound, but as an electron donor, it is difficult for microorganisms to activate. Recently, bacteria affiliated with the “NC10” phylum that couple anaerobic methane oxidation to nitrite reduction (n-damo) were discovered and enriched from different freshwater systems in The Netherlands and Australia and were tentatively named “Candidatus Methylomirabilis oxyfera” (7, 10, 17, 21). If methane could be used to drive denitrification, this would circumvent the need for expensive electron donors (for example, methanol for nitrogen removal in industrial wastewater treatment systems). Coupling of anammox,
and n-damo,
represents untapped potential for wastewater treatment systems. The combination of these processes would be able to remove ammonium, dissolved methane, and nitrite simultaneously without the need for extra aeration and the addition of electron donors.
Besides the availability of ammonium and methane in man-made systems, these compounds are also present in natural anoxic ecosystems. Physiological studies and environmental surveys revealed that both groups of microorganisms may inhabit oxygen-limited ecosystems where methane, ammonium, and nitrogen oxides (NOx) are available (7, 20, 25, 32). Apparently, even though the bacteria that perform these processes compete for the same electron acceptor (nitrite), they are able to coexist in nature.
A wastewater treatment plant is fundamentally different from oxygen-limited ecosystems due to the high volumetric loading rates and nutrient fluxes. Therefore, it is necessary to study the feasibility of cocultures where n-damo and anammox processes are performed by two competing groups of microorganisms. In this study, we investigated whether n-damo and anammox bacteria could coexist under conditions relevant to wastewater treatment plants.
MATERIALS AND METHODS
Enrichment of anaerobic methane- and ammonium-oxidizing bacteria.
A previously described enrichment culture (1 liter) of anaerobic methane-oxidizing bacteria (9) was used to inoculate a 3-liter sequencing batch reactor (SBR) for the enrichment of anammox bacteria. Each SBR cycle consisted of 11 h 45 min of constant medium supply, 5 min of settling for biomass retention, and 10 min of removal of excess liquid. To maintain anoxic conditions and to supply the culture with methane, the SBR was flushed continuously with CH4-CO2 (95:5%, 12 ml min−1). The medium vessel was flushed with Ar-CO2 (95:5%, 10 ml min−1). The SBR was stirred at 200 rpm with a turbine stirrer, and the temperature was kept constant at 30°C. The supplied CO2, together with the bicarbonate in the synthetic medium, kept the pH of the culture between 7.3 and 7.6. During each filling period, synthetic medium (9) was added continuously. The flow rate was adjusted based on the nitrite-reducing activity of the culture. In the first 0 to 49 days of the experiment, no ammonium was added to achieve steady-state conditions. On day 50, ammonium was added to the medium (1 mM NH4+, about 400 ml day−1) to induce the growth of anammox bacteria. Nitrite and ammonium concentrations were further increased during the experiment to keep up with the growth of the microorganisms (see the section on the enrichment of anammox bacteria in Results).
DNA isolation and phylogenetic analysis.
Biomass (2 ml) from the SBR was collected by centrifugation and used for DNA extraction at three different time points: when no ammonium was present in the SBR on day 22 and after the addition of ammonium on days 87 and 139. DNA extraction and purification were performed according to reference 18. To detect n-damo and anammox bacteria, primers targeting the functional gene pmoA and the 16S rRNA gene were used as described previously (9, 18, 24). For the detection of anammox bacteria on day 22, primers Pla46 (19) and 1545R (12) were used in a nested-PCR approach with AMX368F and AMX820R (24). The PCR fragments from the same gradient PCR were mixed and ligated into the pGEM-T Easy cloning vector according to manufacturer's protocol (Promega). Plasmid DNA was isolated and purified with the Gene JET Plasmid Miniprep kit (Fermentas, Vilnius, Lithuania). Sequence analysis and alignment were performed as described previously (18). Alignments were performed using the ClustalW algorithm, and phylogenetic trees were calculated by the neighbor-joining method. Tree topology robustness was tested by bootstrap analysis of 1,000 replicates using the MEGA4 software package (28).
Activity measurements.
To determine the nitrite, ammonium, and methane consumption rates in the SBR, two batch incubations were performed. The first activity measurement was on day 49, when no ammonium was present. The methane flow was stopped, and the headspace of the bioreactor was flushed with Ar-CO2 (10 ml min−1) for 15 min. Subsequently, the influent was stopped and the headspace was closed completely. Methane (50 ml) was added to the headspace, yielding a final concentration of 8% CH4. Nitrite was added to the reactor to a final concentration of 1.3 mM NO2−. After an incubation period of ∼12 h, methane, nitrite, and ammonium samples were taken every hour and analyzed as described below in the section on analytical methods.
In the second batch incubation (day 142), methane was present in excess and the final concentrations of nitrite and ammonium in the SBR were 1.2 mM and 0.5 mM, respectively. Samples were taken for nitrite and ammonium analyses, which were performed as described below in the section on analytical methods.
Fluorescence in situ hybridization (FISH).
Biomass (1.5 ml) was harvested from the enrichment culture, fixed in paraformaldehyde, and hybridized as described previously (23). All probes were purchased as Cy3-, Cy5-, and 5(6)-carboxyfluorescein N-hydroxysuccinimide ester (FLUOS)-labeled derivatives from Thermo Electron Corporation (Ulm, Germany). The following probes were used to monitor the anaerobic ammonium- and methane-oxidizing communities: S-*-Amx-0368-a-A-18 (Amx368), which is specific for all known anammox genera (23); S-*-Amx-0820-a-A-22 (Amx820), which is specific for “Candidatus Kuenenia stuttgartiensis” and “Candidatus Brocadia anammoxidans” (22); EUBmix (i.e., EUB338, EUB338II, and EUB338III mixed in an equimolar solution), which is specific for most bacteria (2, 6); S-P-Planc-0046-a-A-18 (Pla46), which was used to detect Planctomycetales (19); and S-*-DBACT-1027-a-A-18 (DBact1027), which was used for bacteria affiliated with the “NC10” phylum (21). All samples were counterstained with the DNA stain 4′,6-diamidino-2-phenylindole (DAPI). The slides were examined using a Zeiss Axioplan II epifluorescence microscope with a digital video camera and image analysis software (AxioVision; Zeiss, Jena, Germany).
Analytical methods.
Nitrite, nitrate, and ammonium were measured as described in reference 13. For routine nitrite and nitrate analyses, Merckoquant test strips (Merck, Darmstadt, Germany) ranging from 0 to 80 mg/liter and from 0 to 500 mg/liter, respectively, were used. Methane was quantified by gas chromatography, and the total protein content was determined by the bicinchoninic acid assay (Pierce) as described before (8).
Nucleotide sequence accession numbers.
Representative sequences of anammox bacteria (16S rRNA) and n-damo bacteria (pmoA) from every time point were deposited in GenBank under accession numbers JN006723 to JN006737.
RESULTS
Source of inoculum.
An SBR was inoculated on day 0 with 1 liter of an established anaerobic methane-oxidizing culture containing 70 to 80% n-damo bacteria (9). After 49 days of stable operation, an activity test was performed with the whole reactor (0.12 g protein) to determine the specific activity of the n-damo bacteria. The methane and nitrite consumption rates were 3.1 nmol min−1 mg protein−1 and 8.6 nmol min−1 mg protein−1, respectively. The measured ratio of methane to nitrite was 3:8.1, which is identical to the theoretical stoichiometry (see the d-damo process equation in the introduction). The composition of the n-damo culture on day 45 (see Fig. 3A) was visualized by FISH analysis. n-damo cells made up 80% of the community, whereas anammox bacteria were below the limit of detection by FISH (∼10,000 cells/ml) (3).
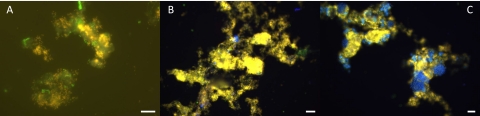
Fig. 3.
In situ detection of n-damo and anammox bacteria using fluorescence probes. n-damo bacteria were identified using Cy3 (yellow) DBACT1027. Anammox bacteria were identified using Cy5 (light blue) AMX368. (A) On day 45, n-damo bacteria were present but no anammox bacteria could be detected. EUBmix was used in FLUOS (green). (B) After 106 days, anammox cells were growing in clusters together with the n-damo bacteria. (C) On day 151, the anammox and n-damo populations were equally distributed (50% of each group of organisms). Scale bars: 10 μm.
DNA was extracted from the n-damo enrichment culture, and clone libraries for the pmoA gene of these bacteria were subsequently constructed (day 22). Clones of the pmoA gene were closely related (nucleotide sequence identities ranged from 91 to 95%) to the pmoA gene found in the published genome of “Candidatus Methylomirabilis oxyfera” (Fig. 1A). It was not possible to detect the anammox bacteria in the enrichment culture by a direct PCR approach. Therefore, a nested PCR using primers targeting the 16S rRNA gene of anammox bacteria was performed to verify if these bacteria were present besides the dominant n-damo bacteria. This resulted in clones that were similar to the known anammox bacteria “Candidatus Jettenia asiatica,” “Candidatus Brocadia fulgida,” and “Candidatus Kuenenia stuttgartiensis” (Fig. 1B). After confirmation of the presence of residual anammox bacteria in the n-damo enrichment culture, addition of ammonium was started in order to obtain a coculture.
Fig. 1.
Phylogenetic trees of pmoA sequences (n-damo) and 16S rRNA sequences (anammox). The clone libraries are named after the time points when DNA was extracted, i.e., at 22 (when no ammonium was present), 87, and 139 days. Trees were calculated by the neighbor-joining method, and tree topology robustness was tested by bootstrap analysis with 1,000 replicates. Bootstrap support values of greater than 70% are indicated at the nodes. Nucleotide sequences of amoA, pmoA, pxmA (A), and anammox 16S rRNA (B) are represented, including the sequences obtained in this study. Scale bars: 0.1 (A) and 0.02 (B) substitution per site.
Enrichment of anammox bacteria.
Medium containing 1 mM ammonium (flow, about 400 ml day−1) was supplied to the n-damo enrichment culture at day 50. Significant ammonium consumption (0.2 mmol day−1) was measured within 14 days (Fig. 2A). From day 80, the ammonium concentration in the medium was gradually increased to 20 mM, in proportion with the ammonium consumption (Fig. 2A). To provide anammox and n-damo bacteria with enough nitrite, the concentration of nitrite in the medium was increased to 37.5 mM. Nitrate production was in line with the increasing nitrite and ammonium consumption by anammox bacteria. Methane and nitrite were always supplied to the reactor in excess, whereas the anammox bacteria were always kept under ammonium limitation. In this way, an actively growing coculture of anammox and n-damo bacteria was established.
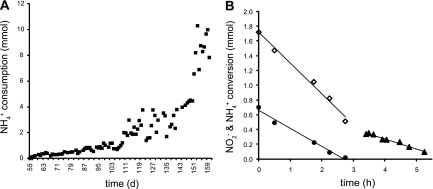
Fig. 2.
Increasing rates of NH4+ consumption by the whole culture after the addition of ammonium to an enrichment culture performing nitrite-dependent anaerobic methane oxidation. (A) On day 50, ammonium was added to the medium. Consumption was measured on day 55. The ammonium concentrations in the medium could be increased to 20 mM at day 160 (about 400 ml day−1). (B) To determine the proportions of nitrite consumption by anammox and n-damo bacteria, the conversion rates of the coculture were measured on day 142. When nitrite and ammonium were both present (day 0 to 2.8 h), the total nitrite consumption was 15.4 nmol min−1 mg protein−1 (open diamonds) and the ammonium consumption was 9.0 nmol min−1 mg protein−1 (closed circles). When ammonium was depleted, nitrite was consumed at 5.0 nmol min−1 mg protein−1 (closed triangles).
Specific activities of the coculture.
To determine the contributions of n-damo and anammox bacteria to the consumption of nitrite, an activity test with the whole enrichment culture (0.45 g protein) was performed on day 142 (Fig. 2B). Nitrite and ammonium consumption was determined for 6 h. Within the first 3 h, nitrite was converted at a rate of 15.4 nmol min−1 mg protein−1 and ammonium was converted at a rate of 9.0 nmol min−1 mg protein−1 (Fig. 2B). In this period, the anammox and n-damo processes were occurring simultaneously. Using the anammox stoichiometry of 1:1.32 (ammonium to nitrite) (27), the theoretical nitrite reduction rate of anammox bacteria was calculated from the ammonium concentration as 11.9 nmol min−1 mg protein−1 in the first 3 h of incubation. After 3 h, ammonium was depleted and nitrite was converted with 5.0 nmol min−1 mg protein−1.
Identification of anammox and n-damo bacteria in the coculture.
The pmoA clones (days 87 and 139) for the detection of n-damo cells showed high similarity (nucleotide sequence identities ranged from 89 to 99.7%) to the clones obtained on day 22 and “Candidatus Methylomirabilis oxyfera” (Fig. 1A). These samples were also screened with 16S rRNA primers specific for n-damo bacteria and showed that all of the clones obtained clustered within group a of the “NC10” phylum (9; data not shown). There was an apparent shift in the anammox population (day 87 and 139) compared to the original sample (day 22); bacterial 16S rRNA sequences related to “Candidatus Kuenenia stuttgartiensis” were no longer detected, but 13 (out of 13) clones at day 139 showed similarity to either “Candidatus Jettenia asiatica” or “Candidatus Brocadia fulgida” (Fig. 1B).
The coculture community composition was monitored by FISH. On day 106, anammox cells were visible and represented approximately 5% of the community (Fig. 3B). Anammox cells were growing in clusters, and together with n-damo bacteria, they formed flocks. After 151 days, the coculture constituted about 50% of each group of organisms (Fig. 3C).
DISCUSSION
Anammox bacteria were enriched using an existing n-damo culture as the inoculum in order to study the possibility of simultaneous ammonium and methane removal by these two groups of microorganisms. At the start of the experiments, when no ammonium was present, the n-damo activity was approximately 1.7 times higher than that previously reported (9). At that time, sequences of n-damo and anammox bacteria were detected. The detection of anammox bacteria on day 22 indicated that these bacteria had persisted for at least 8 months of cultivation without external ammonium supply to the culture. Mineralization of organic matter and biomass decay might have supplied anammox bacteria with very small amounts of ammonium, while sufficient nitrite was supplied with the synthetic medium. The sequences obtained after 139 days showed a shift in the anammox population. In previous studies, it was shown that the addition of organic acids could cause a shift in the anammox community (15, 16). It is possible that in the present coculture, organic compounds released through biomass decay or the presence of methane led to a shift toward the species “Candidatus Brocadia fulgida” and “Candidatus Jettenia asiatica”. On the other hand, pmoA clones from the enriched coculture (days 89 and 139) formed separate clusters within the sequences belonging to the “NC10” phylum, suggesting that there was a degree of microdiversity in the n-damo population, as was also observed in various wastewater treatment plants (17).
After the coculture was established, the n-damo activity was on the same order as previously reported (8, 9, 21). Methane and nitrite were always present in excess, while the concentration of ammonium was limited. Nevertheless, anammox bacteria were responsible for 77% of the nitrite consumption after 142 days of cultivation, when they constituted 50% of the coculture. Most likely, if ammonium was in excess, anammox bacteria would probably outcompete n-damo bacteria (11), suggesting that anammox bacteria have a higher affinity for nitrite.
In current full-scale anammox bioreactors, nitrite is supplied through partial nitrification by aerobic ammonium-oxidizing bacteria (1). In these systems, such as the completely autotrophic nitrogen removal over nitrite process, aerobic and anaerobic ammonium-oxidizing microorganisms convert inorganic nitrogen compounds under oxygen-limited conditions in a single-stage reactor (26, 29, 31). In similar oxygen-limited systems, the n-damo bacteria would have to compete with both aerobic methane-oxidizing and anaerobic ammonium-oxidizing bacteria. Additional experiments with laboratory scale bioreactors and feasibility tests with pilot scale wastewater treatment plants are necessary to test the competitive fitness of n-damo bacteria under conditions relevant to wastewater treatment to determine the applicability of the coculture described here.
ACKNOWLEDGMENTS
This study was supported by the Microbiology Department of Radboud University Nijmegen; the Universidad de los Andes, Mérida, Venezuela; and Universidad del Valle, Cali, Colombia.
We thank Katharina Ettwig, Jan Keltjens, and Katinka van de Pas-Schoonen for discussions, Mathilde le Roy and Dirk Verhijen for initial screening of anammox 16S rRNA genes, and Elisabeth Pierson for confocal microscopy. F.A.L. and T.A.V.A. were supported by the Foundation for Applied Research (STW project 07736). M.S.M.J. was supported by ERC grant 2322937. B.K. is supported by KRW grant 09035.
Footnotes
Published ahead of print on 12 August 2011.
Both authors contributed equally.
REFERENCES
- 1. Abma W. R., Driessen W., Haarhuis R., van Loosdrecht M. C. M. 2010. Upgrading of sewage treatment plant by sustainable and cost-effective separate treatment of industrial wastewater. Water Sci. Technol. 61:1715–1722 [DOI] [PubMed] [Google Scholar]
- 2. Amann R. I., et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amann R. I., Ludwig W., Schleifer K. H. 1995. Phylogenetic identification and in-situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bogner J., et al. 2008. Mitigation of global greenhouse gas emissions from waste: conclusions and strategies from the Intergovernmental Panel on Climate Change (IPCC) fourth assessment report. Working Group III (Mitigation). Waste Manag. Res. 26:11–32 [DOI] [PubMed] [Google Scholar]
- 5. Cakir F. Y., Stenstrom M. K. 2005. Greenhouse gas production: a comparison between aerobic and anaerobic wastewater treatment technology. Water Res. 39:4197–4203 [DOI] [PubMed] [Google Scholar]
- 6. Daims H., Bruhl A., Amann R., Schleifer K. H., Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 7. Ettwig K. F., et al. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548 [DOI] [PubMed] [Google Scholar]
- 8. Ettwig K. F., et al. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10:3164–3173 [DOI] [PubMed] [Google Scholar]
- 9. Ettwig K. F., van Alen T., T. van de Pas-Schoonen K., Jetten M. S., Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75:3656–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu S., et al. 2009. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ. Microbiol. Rep. 1:377–384 [DOI] [PubMed] [Google Scholar]
- 11. Jetten M. S. M., Op den Camp H. J. M., Keltjens J. T., Strous M. 2008. Two impossible microbes with global implications: the nitrite-dependent anaerobic oxidation of methane and ammonium, p. 15–22.In Liu S. J., Drake H. L. (ed.), Microbes and the environment: perspective and challenges. Science Press, Beijing, China. [Google Scholar]
- 12. Juretschko S., et al. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kartal B., et al. 2006. Adaptation of a freshwater anammox population to high salinity wastewater. J. Biotechnol. 126:546–553 [DOI] [PubMed] [Google Scholar]
- 14. Kartal B., Kuenen J. G., van Loosdrecht M. C. M. 2010. Sewage treatment with anammox. Science 328:702–703 [DOI] [PubMed] [Google Scholar]
- 15. Kartal B., et al. 2007. Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 30:39–49 [DOI] [PubMed] [Google Scholar]
- 16. Kartal B., et al. 2008. Candidatus ‘Brocadia fulgida': an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol. Ecol. 63:46–55 [DOI] [PubMed] [Google Scholar]
- 17. Luesken F. A., et al. 11 June 2011, posting date. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl. Microbiol. Biotechnol. [Epub ahead of print.] doi:10.1007/s00253-011-3361-9. [DOI] [PMC free article] [PubMed]
- 18. Luesken F. A., et al. 2011. pmoA primers for detection of anaerobic methanotrophs. Appl. Environ. Microbiol. 77:3877–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neef A., Amann R., Schlesner H., Schleifer K. H. 1998. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144(Pt. 12):3257–3266 [DOI] [PubMed] [Google Scholar]
- 20. Penton C. R., Devol A. H., Tiedje J. M. 2006. Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl. Environ. Microbiol. 72:6829–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raghoebarsing A. A., et al. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921 [DOI] [PubMed] [Google Scholar]
- 22. Schmid M., et al. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93–106 [DOI] [PubMed] [Google Scholar]
- 23. Schmid M., et al. 2003. Candidatus “Scalindua brodae,” sp nov., Candidatus “Scalindua wagneri,” sp nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26:529–538 [DOI] [PubMed] [Google Scholar]
- 24. Schmid M. C., et al. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 71:1677–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmid M. C., et al. 2007. Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ. Microbiol. 9:1476–1484 [DOI] [PubMed] [Google Scholar]
- 26. Sliekers A. O., et al. 2002. Completely autotrophic nitrogen removal over nitrite in one single reactor. Water Res. 36:2475–2482 [DOI] [PubMed] [Google Scholar]
- 27. Strous M., Heijnen J. J., Kuenen J. G., Jetten M. S. M. 1998. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 50:589–596 [Google Scholar]
- 28. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 29. Third K. A., Sliekers A. O., Kuenen J. G., Jetten M. S. M. 2001. The CANON system (completely autotrophic nitrogen-removal over nitrite) under ammonium limitation: interaction and competition between three groups of bacteria. Syst. Appl. Microbiol. 24:588–596 [DOI] [PubMed] [Google Scholar]
- 30. van der Star W. R., et al. 2007. Startup of reactors for anoxic ammonium oxidation: experiences from the first full-scale anammox reactor in Rotterdam. Water Res. 41:4149–4163(Errata, 42:1825-1826 and 44:1025) [DOI] [PubMed] [Google Scholar]
- 31. Yan J., Op den Camp H. J., Jetten M. S., Hu Y. Y., Haaijer S. C. 2010. Induced cooperation between marine nitrifiers and anaerobic ammonium-oxidizing bacteria by incremental exposure to oxygen. Syst. Appl. Microbiol. 33:407–415 [DOI] [PubMed] [Google Scholar]
- 32. Zhu G. B., Jetten M. S. M., Kuschk P., Ettwig K. F., Yin C. Q. 2010. Potential roles of anaerobic ammonium and methane oxidation in the nitrogen cycle of wetland ecosystems. Appl. Microbiol. Biotechnol. 86:1043–1055 [DOI] [PubMed] [Google Scholar]