Abstract
Considerable research has shown that cyanobacteria and the viruses that infect them (cyanophage) are pervasive and diverse in global lake populations. Few studies have seasonally analyzed freshwater systems, and little is known about the bacterial and viral communities that coexist during the harsh winters of the Laurentian Great Lakes. Here, we employed quantitative PCR to estimate the abundance of cyanomyoviruses in this system, using the portal vertex g20 gene as a proxy for cyanophage abundance and to determine the potential ecological relevance of these viruses. Cyanomyoviruses were abundant in both the summer and the winter observations, with up to 3.1 × 106 copies of g20 genes ml−1 found at several stations and depths in both seasons, representing up to 4.6% of the total virus community. Lake Erie was productive during both our observations, with high chlorophyll a concentrations in the summer (up to 10.3 μg liter−1) and winter (up to 5.2 μg liter−1). Both bacterial and viral abundances were significantly higher during the summer than during the winter (P < 0.05). Summer bacterial abundances ranged from 3.3 × 106 to 1.6 × 107 ml−1 while winter abundances ranged between ∼3.4 × 105 and 1.2 × 106 ml−1. Total virus abundances were high during both months, with summer abundances significantly higher at most stations, ranging from 6.5 × 107 to 8.8 × 107 ml−1, and with winter abundances ranging from 3.4 × 107 to 6.6 × 107 ml−1. This work confirms that putative cyanomyoviruses are ubiquitous in both summer and winter months in this large freshwater lake system and that they are an abundant component of the virioplankton group.
INTRODUCTION
Since the late 1980s, research on aquatic viruses has increased dramatically as the collective community of microbial ecologists has realized the important roles that viruses may play in aquatic ecosystems (4, 41). Despite the wealth of information that has been produced in marine ecosystems, published research on viruses in freshwater environments remains sparse. To date, researchers have shown that viruses in freshwater environments are ubiquitous and often, although not always (7), persist at higher densities than do their marine counterparts (e.g., see reference 8). Viruses in marine environments have been linked to the biogeochemical cycling of nutrients (P, N, and Fe) as well as carbon, but only limited work has been done to confirm this in freshwater systems (50).
Cyanobacteria, including Synechococcus (16, 47), Microcystis (28, 29), and Planktothrix (31) spp., have been shown to be prevalent in different parts of Lake Erie and are potential hosts for cyanophage infection. To this end, researchers have developed primers specific for genes within groups of viruses, such as the g20 gene used in the study of Cyanomyoviridae (35). The g20 gene encodes a T4 portal-protein homologue in Cyanomyoviridae and has been used for studying the phylogenetic diversity of the phage infecting cyanobacteria in both marine (21, 25, 55) and freshwater (9, 33, 48) ecosystems. While significant genetic richness in both environments has been evident, little has been done to quantify these virus particles.
Approximately 20% of the world's freshwater is contained within North America's Laurentian Great Lakes (45). Lake Erie is the smallest (by volume), shallowest, and most productive of the Laurentian Great Lakes. Despite this, it is the 18th largest freshwater system in terms of volume (483 km3). The lake's large population base of over 30 million people, in addition to agricultural and industrial influences, has resulted in historically high biological productivity in this lake and subsequently ecosystem degradation (26). Excessive primary productivity has been the highlight of research for decades in this system; most recently, studies have focused on Lake Erie due to seasonal harmful algal blooms that form during the summer months (1, 3, 30). Factors promoting or constraining primary production in the lake are thus of great interest not only to the academic community but also to ecosystem managers.
Cyanophage abundance has historically been estimated via infection assays such as the most probable number method (12). One problem with this approach is that this and other assays are constrained to viruses that infect laboratory isolates of cyanobacteria: these assays, as such, likely miss a significant proportion of cyanophage that may not infect (or may not infect under the chosen laboratory conditions) the model cyanobacterium chosen. To better understand the role that cyanomyoviruses infecting Synechococcus spp. may play in freshwater ecosystems, we set out to develop quantitative estimates of their abundance by employing the g20 gene as a molecular proxy for the Cyanomyoviridae. Moreover, to begin to address seasonal alterations in virus density in the water column, samples from Lake Erie were collected during both winter (February) and summer (August) expeditions in 2009. To our knowledge, only two papers have been published looking at the abundances of cyanophage using a quantitative PCR (qPCR) approach, with one focusing on viruses near a coastal marine station (32) and the second targeting viruses specific for Microcystis (38). Our results demonstrate that this marker gene (and thus putative cyanomyoviruses) is ubiquitous and abundant across Lake Erie during both the summer and winter months and that there are temporal changes in both bacteria and viruses in this system. When taken together with other metadata, our observations reinforce the potential ecological importance of virus infection of cyanobacteria in this important freshwater resource.
MATERIALS AND METHODS
Sampling.
Winter water samples were collected during daylight hours from Lake Erie using the icebreaker CCGS Griffon as a research platform from 16 to 20 February 2009. Where ice cover was evident, small patches of ice were cleared with the ship (and debris was subsequently allowed to settle for >30 min). Samples were collected at identical locations in both winter and summer using Niskin bottles, which in the summer were attached to the ship's rosette sampling system. To record the structure of the water column and determine the depths of interest for sampling, we used a YSI Model 6600 Sonde (YSI Instruments, Yellow Springs, OH) on the winter cruise to obtain depth profiles of temperature, pH, specific conductance, dissolved oxygen, turbidity, and fluorescence (as a proxy for chlorophyll a [Chl a]). On the summer cruise, temperature, pH, specific conductance, dissolved oxygen, and light transmission (a proxy for turbidity) were profiled using a Sea-Bird Model 25 instrument (Sea-Bird Electronics, Inc., Belleview, WA) and fluorescence profiles were obtained with a custom system incorporating a Seapoint chlorophyll fluorometer (Seapoint Sensors, Inc., Exeter, NH).
Measurement of nutrients and chlorophyll a.
The concentrations of nutrients (NO3−, NH3, SiO2, soluble reactive phosphorus [SRP], total dissolved nitrogen, and total dissolved phosphorus) were measured from water samples filtered on board through 0.45-μm cellulose acetate membranes and stored at 4°C until analysis was completed at the National Laboratory for Environmental Testing (Environment Canada) using standardized techniques (49).
Size-fractionated Chl a concentrations were determined on duplicate samples collected on 0.22- (for picophytoplankton)-, 2.0- (for nanophytoplankton)-, and 20.0-μm (for microphytoplankton)-nominal-pore-size, 47-mm-diameter polycarbonate filters (GE Osmonics). Samples were extracted in 90% acetone for 24 h at 4°C, and Chl a retained on the filters was quantified using a solid-standard normalized 10-AU field fluorometer (Turner Designs) using the nonacidification protocol (44).
Virus abundance.
The abundance of virus particles was determined for whole-water samples which were preserved in 2.5% (final) sterile glutaraldehyde. Abundance samples for the winter Lake Erie stations were preserved and stored at 4°C until processed (<2 weeks), while summer samples were flash frozen in liquid nitrogen and stored at −20°C until analysis. Slide preparation proceeded as previously described for 850 μl of thawed sample (27a). All slides were stored at −20°C until virus-like particles could be enumerated via epifluorescence microscopy using a Leica DMRXA with a “wide blue” filter set (λEx = 450 to 490 nm; λEm = 510 nm). At least 20 independent fields or 200 particles were examined, with the total of each field of view noted to ensure even distribution of particles across individual filters, for each slide.
Bacterial and cyanobacterial abundance.
Samples collected for bacterial abundance estimates (including Bacteria as well as Archaea) during Lake Erie cruises were suspended in 2.5% (final) sterile glutaraldehyde and kept at 4°C in the dark until processed in the laboratory. Fixed samples were stained for 3 min with a 1% working solution of acridine orange stain (14). The stained cells were then collected on a 0.22-μm-nominal-pore-size, 25-mm-diameter black polycarbonate filter (GE Osmonics), and cells were counted as described above for viral abundances. Synechococcus and cyanobacterial abundances were estimated using the Texas Red filter set (λEx, 595 nm; λEm, 610 to 615 nm) on the same slides as those used for bacterial counts.
Quantitative PCR of g20 gene.
Samples for qPCR analysis were collected in 5-ml cryovials, immediately flash frozen in liquid N2, and stored at −80°C until they were processed. The primer set CPS1/CPS2 was used to estimate g20 gene copies (11). Plasmid standards were prepared using Synechococcus phage S-PWM1; the g20 gene was amplified using primer pair CPS1/CPS8 (55), which produces a 592-bp product that contains the 165-bp fragment from CPS1 and CPS2. PCR with S-PWM1 phage was completed with PuReTaq Ready-To-Go PCR beads (GE Healthcare, Buckinghamshire, United Kingdom) using 0.2 μM (each) primer, 1 μl of filtered S-PWM1 phage stock, and sterile water and reaction conditions previously reported (55). The DNA fragment was cloned into the PCR 2.1 TOPO-TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Plasmid DNA was purified using the Qiagen Qiaprep Spin Miniprep kit (Qiagen, Valencia, CA), and cloned inserts were confirmed using an EcoRI digestion at 37°C for 2 h followed by electrophoresis with 1.5% agarose gels. The plasmid DNA sequence was confirmed at the University of Tennessee Sequencing Facility. The DNA concentration (A260) of the plasmid was determined spectrophotometrically using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Surrey, United Kingdom). The molecular weights of the double-stranded plasmid and product were calculated using the Oligocalc Calculator version 3.23 (17). Copy numbers of g20 per μl were determined using Avogadro's number (6.022 × 1023 plasmid copies mol−1). Each plasmid contained one copy of the target gene and was freshly diluted and used within 24 with at least four different duplicate dilutions from 106 to 101 copies to make a linear standard curve for the analyses. Each 25-μl qPCR mixture contained 12.5 μl Thermo Scientific ABsolute SYBR green qPCR mix (Thermo Scientific, Surrey, United Kingdom), 280 ng μl−1 bovine serum albumin (Fisher Scientific), 0.30 μM CPS1 forward primer and 0.60 μM CPS2 reverse primer, and 2 μl viral sample or standard.
Environmental samples were ultracentrifuged at 30,000 to 35,000 rpm (Beckman TL-100) for 3 h at 4°C and kept at −20°C prior to direct qPCR. Samples were run in triplicate and assayed with up to three different dilutions. PCR conditions consisted of an initial denaturation step at 95°C for 15 min followed by 35 cycles of 95°C for 10 s, 53°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min. Following the program, a melting curve was read every 1°C from 40 to 95°C to check specificity of primers and formation of primer dimers. As nonspecific binding was exhibited at the lowest copy numbers, a second plate reading at 77°C was performed after every cycle to inhibit the fluorescence detection of primer dimers (5, 24). Negative control assays were conducted along with samples using sterile water and no DNA template. Positive controls consisted of lysate from Synechococcus phage S-PWM1. To determine the potential contribution of viruses within cells relative to free viruses, winter samples were passed through a 1-μm-nominal-pore-size 47-mm polycarbonate filter on a Sterifilter unit and compared to unfiltered and untreated samples.
Virus contact rates.
Contact rates of putative cyanomyoviruses (from g20 gene copy numbers) with total bacterial populations and cyanobacteria were estimated following the equation from Murray and Jackson (27), (2 · S · π · ω · Dv)V · B, using the dimensionless Sherwood number (S) of 1.06 for a 10% motile population (52). The average diameter (ω) of a bacterial cell (1.0 × 10−4 cm) (10) and the diffusivity of viruses (Dv) (3.456 × 10−3 cm2 day−l) were taken from previous sources (27). Viral abundance (V) and bacterial abundance (B) were both expressed per ml−1. For contact rates between viruses or cyanophage and cyanobacterial cells, a diameter of 1.5 × 10−4 cm was used for Synechococcus (36). Specific contact rates of viruses per bacterium per day were determined by dividing the contact rates by the host abundance.
Statistical analyses.
Threshold cycle (CT) calculations were made for each qPCR assay using the MJ Opticon Monitor Analysis software (version 3.1) with the Global Minimum setting. The threshold was manually adjusted for each run to give the highest correlation coefficient (r2) for each standard curve. Gene copy abundance in each sample was calculated based on the standard curve (gene copy number versus CT). Averages, standard deviations, F tests (for variance), and paired and unpaired two-tailed t tests were determined using Microsoft Excel 2007. Pearson correlations for temperature, latitude, virus abundances, bacterial abundances, g20 gene copies, cyanobacterial abundance, and Chl a were analyzed using the NCSS Statistical and Power Analysis software.
RESULTS
Water conditions.
Samples were collected in February and August of 2009 at several stations in the Lake Erie basin (Fig. 1). Lake Erie was isothermal during the winter with water temperatures at or near freezing at all stations and depths (Table 1). Winter temperatures ranged from 0.0 to 0.4°C, with the highest temperatures at the snow- and ice-covered stations 341 and 357. Summer temperatures in the lake were warmest at the surface (24.0 to 25.1°C) and decreased with depth (11.9 to 24.9°C). Stations analyzed during the summer were thermally stratified with the exception of station 357, which seasonally remains destabilized due to its shallow depth.
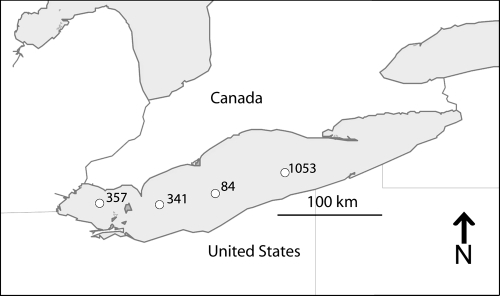
Fig. 1.
Four Lake Erie stations where samples were collected in February and August 2009 (station 84, 41°56.08′N, 80°52.18′W; station 1053, 42°10.54′N, 80°52.18′W; station 357, 41°49.30′N, 82°58.30′W; station 341, 41°48.00′N, 82°17.30′W).
Table 1.
Water column characteristics during sample collectiona
| Station | Depth (m) | Date (mo/day) | Concn (mg liter−1) of nutrient or DO |
Temp (°C) | Chl a concn, μg liter−1 (SD, n = 2) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NO3 | NH3 | SiO2 | SRP | TN | TP | DO | 0.2–2 μm | 2–20 μm | <20 μm | ||||||||
| 84 | 1 | 2/19 | 0.15 | 0.016 | 1.18 | 0.0098 | 0.390 | 0.013 | 17.34 | 0.03 | 0.66 (0.15) | 0.84 (0.03) | 0.67 (0.01) | ||||
| 1 | 8/18 | 0.02 | 0.009 | 0.3 | 0.0007 | 0.288 | 0.076 | 9.13 | 24.63 | 5.08 (0.11) | 3.15 (0.11) | 0.65 (0.00) | |||||
| 20 | 2/19 | 0.14 | 0.011 | 1.18 | 0.0085 | 0.396 | 0.011 | 17.23 | 0.03 | 0.62 (0.07) | 0.75 (0.09) | 0.65 (0.04) | |||||
| 18 | 8/18 | 0.11 | 0.064 | 0.2 | 0.0010 | 0.391 | 0.007 | 7.44 | 22.46 | 0.7175 (0.00) | 0.32 (0.02) | 0.19 (0.00) | |||||
| 1053 | 1 | 2/19 | 0.17 | 0.007 | 0.81 | 0.0069 | 0.393 | 0.011 | 15.54 | 0.06 | 2.21 (0.41) | 2.55 (0.35) | 1.97 (0.03) | ||||
| 1 | 8/19 | 0.09 | 0.039 | 0.21 | 0.0011 | 0.308 | 0.007 | 9.69 | 25.1 | 2.84 (0.69) | 2.14 (0.01) | 0.19 (0.06) | |||||
| 18 | 2/19 | 0.16 | 0.011 | 0.84 | 0.0080 | 0.436 | 0.011 | 15.46 | 0.04 | 2.17 (0.07) | 2.61 (0.18) | 2.02 (0.00) | |||||
| 18 | 8/19 | 0.09 | 0.041 | 0.41 | 0.0013 | 0.318 | 0.007 | 8.39 | 21.75 | 0.7268 (0.44) | 0.64 (0.02) | 0.23 (0.03) | |||||
| 357 | 1 | 2/17 | 0.42 | 0.025 | 1.99 | 0.0043 | 0.665 | 0.006 | 15.32 | 0.39 | 0.40 (0.02) | 0.15 (0.02) | 0.03 (0.01) | ||||
| 1 | 8/18 | 0.07 | 0.042 | 0.82 | 0.0013 | 0.330 | 0.007 | 7.93 | 24.9 | 6.955 (3.25) | 6.44 (2.14) | 6.08 (0.79) | |||||
| 8 | 2/17 | 0.44 | 0.027 | 1.99 | 0.0041 | 0.612 | 0.005 | 14.97 | 0.42 | 0.35 (0.02) | 0.17 (0.00) | 0.04 (0.00) | |||||
| 8 | 8/18 | 0.07 | 0.051 | 0.83 | 0.0013 | 0.335 | 0.007 | 9.08 | 24.88 | 10.31 (1.12) | 6.08 (0.46) | 4.85 (0.18) | |||||
| 341 | 1 | 2/17 | 0.40 | 0.035 | 1.56 | 0.0081 | 0.634 | 0.010 | 16.16 | 0.39 | 3.54 (0.00) | 3.44 (0.00) | 2.39 (0.18) | ||||
| 1 | 8/20 | 0.09 | 0.075 | 1.15 | 0.0023 | 0.346 | 0.009 | 7.35 | 23.96 | 5.26 (1.39) | 4.22 (0.39) | 1.94 (0.02) | |||||
| 16 | 2/17 | 0.24 | 0.020 | 0.88 | 0.0055 | 0.456 | 0.007 | 16.27 | 0.39 | 5.19 (0.38) | 6.35 (0.00) | 4.79 (0.55) | |||||
| 15 | 8/20 | 0.10 | 0.061 | 4.46 | 0.0048 | 0.338 | 0.018 | 0.58 | 11.9 | 1.48 (0.02) | 1.10 (0.26) | 0.54 (0.04) | |||||
Abbreviations: SRP, soluble reactive phosphorus; TN, total dissolved nitrogen; TP, total dissolved phosphorus; DO, dissolved oxygen. For NO3 and NH3, results are reported as mg of N/liter.
Nutrient concentrations were generally higher during winter sampling than during the summer (Table 1). No trends in nutrients comparing surface and deep samples could be discerned. NO3− was the significant nitrogen source (relative to NH4+) at all depths. Silica (SiO2) concentrations were higher in the winter (up to 4.46 mg liter−1), which likely supports the abundant diatom community that has been observed (M. R. Twiss, R. M. L. McKay, and S. W. Wilhelm, unpublished data). Dissolved oxygen concentrations were supersaturated in the winter at all stations and depths (Table 1). Seasonal hypoxic conditions were detected in the summer hypolimnetic samples from station 341 and station 84.
The biomass of primary producers was relatively high during both sampling events. Chlorophyll a concentrations were generally higher during summer sampling times, although deep samples from two stations (station 1053 and station 341) contained total chlorophyll a concentrations up to 10 μg liter−1 during the winter. Synechococcus abundance in the summer was estimated to be between 105 and 106 cells ml−1 at all stations, with the highest densities at central basin station 84 both at the surface and at depth. Between the winter and summer, a phytoplankton community shift from a diatom-dominated system is reflected in chlorophyll size distributions as an increase in the nanophytoplankton (Table 1). Total bacterial abundance was significantly higher (P < 0.05) in the summer than in the winter, with abundances of bacteria up to 1 order of magnitude higher found at all stations and depths.
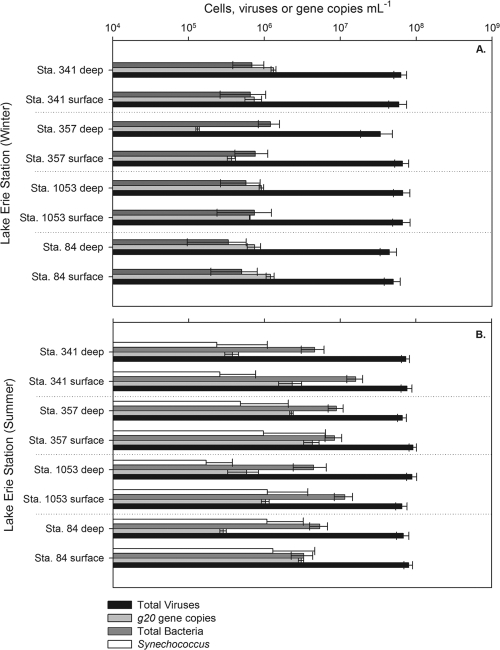
Although we observed shifts in phytoplankton and bacterial biomass, the abundance of viruses at each station remained relatively constant, with slight increases in the summer over the winter (Fig. 2). Total virus particles ranged from 3.4 × 107 to 6.6 × 107 in the winter to 6.5 × 107 to 8.9 × 107 ml−1 in the summer. Statistical analysis showed significantly (P < 0.05) higher viral densities in the summer at all stations examined except for station 1053 at 1 m (P = 0.053). The total virus-to-bacterium ratio (VBR) in the summer ranged from 5.6 to 24.1 viruses per bacterium and from 27.6 to 130.6 in the winter (data not shown).
Fig. 2.
Abundances of total viruses (black), total bacteria (dark gray), Synechococcus (white), and g20 genes (light gray) for each station and month sampled. (A) Abundances in the winter samples. (B) Abundances in the summer samples. Error bars for virus and bacterial abundances were calculated from standard deviations from 20 grid views while g20 gene abundance error bars were calculated from triplicate qPCRs.
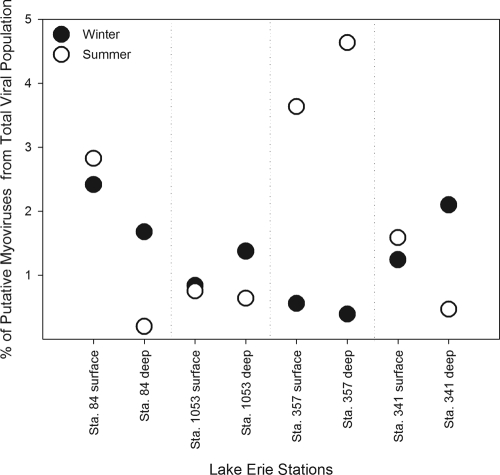
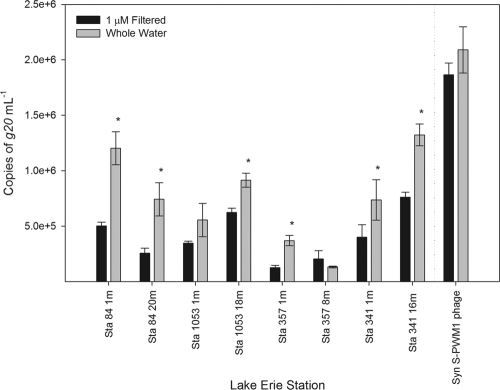
Quantitative PCR targeting the g20 gene estimates for putative cyanomyovirus abundance varied between stations, depth, and season across the range of 1.3 × 105 to 4.3 × 106 ml−1 (Fig. 2). Overall, summer abundances were significantly higher for stations 84 at 1 m (P = 0.002), 1053 at 1 m (P = 0.001), 357 at 1 m (P = 0.02), and 357 at 8 m (P = 0.001) but lower at depth with stations 84 at 18 to 20 m (P = 0.03) and 341 at 15 to 16 m (P = 0.0002). Differences in g20 copies between summer and winter samples for station 1053 at 18 m (P = 0.09) and station 341 at 1 m (P = 0.08) were less significant. The g20-to-bacterium ratio was from 0.11 to 2.37 in the winter with the g20-to-cyanobacterium ratio ranging from 0.26 to 9.02 in the summer sampling. Putative cyanomyoviruses composed from 0.39 to 2.42% of the total viral population in the winter and 0.20 to 4.64% in summer samples (Fig. 3). To determine whether viruses were associated with particulates (i.e., inside or on potential host cells), winter samples were passed through a 1-μm-nominal-pore-size filter and compared to the whole-water samples. A lysate of Synechococcus S-PWM1 phage was also filtered and compared for the analysis. Significant decreases were found at all stations except for station 1053 at 1 m and station 357 at 8 m after filtering the samples, but no significant difference was detected after filtering the phage lysate (Fig. 4).
Fig. 3.
Percentage of the total virus population that is composed of putative cyanomyoviruses using g20 gene copies as a proxy for myovirus abundance. Winter samples are designated by black circles, while summer percentages are designated by white circles.
Fig. 4.
Comparison of Lake Erie winter cyanomyovirus abundance in whole water (black bars) and water that is 1 μm filtered (gray bars). This was compared to a lysate filtered and unfiltered using Synechococcus phage S-PWM1. Error bars represent the standard deviations of triplicate samples from qPCR analysis. Samples that were significantly different (P < 0.05) are designated with an asterisk.
At station 341, hypoxic conditions were detected during summer sample collection at 15 m, with dissolved oxygen concentrations estimated at 0.58 mg liter−1, while concentrations at the surface were 7.35 mg liter−1. Bacterial abundances were significantly higher in the oxic surface waters than in their hypoxic counterpart (P = 0.000), while there was no significant change in virus abundance (P = 0.41) or Synechococcus abundance (P = 0.82).
Contact rates.
Contact rates varied widely with season, station, and depth. Overall, the total contact rates (data not shown) of bacteria to g20 genes ranged from 3.56 × 106 to 8.64 × 107 contacts ml−1 h−1. Total contacts in the summer between cyanobacteria and cyanomyoviruses were estimated and ranged between 3.07 × 105 and 1.44 × 107 (average of 4.92 × 106) contacts ml−1 h−1. Unfortunately, samples for cyanobacterial abundance estimates were lost for the winter months of this sampling period.
To make contact rate information easier to understand and compare, we also calculated cell-specific contact rates for both host (total bacteria, Synechococcus) and virus (total virus particles, putative cyanomyophage) pools (Table 2). Cell-specific contact rates between the total viruses and total bacterial communities ranged from 77.3 to 152.8 (average of 128.9) contacts cell−1 day−1 in the winter and 150.1 to 204.3 (average of 169.4) contacts cell−1 day−1 within the summer.
Table 2.
Contact rates between total viruses or putative cyanophage (g20 copies) and total bacteria or cyanobacteriaa
| Station and season | Depth (m) | Specific contact rate (cell−1 day−1) |
|||
|---|---|---|---|---|---|
| Total viruses (102) |
g20 genes |
||||
| TB | Cyanos | TB | Cyanos | ||
| 84 | |||||
| Winter | 1 | 1.15 | ND | 2.76 | ND |
| 20 | 1.02 | ND | 1.71 | ND | |
| Summer | 1 | 1.84 | 2.76 | 7.01 | 10.52 |
| 18 | 1.56 | 2.35 | 0.66 | 0.98 | |
| 1053 | |||||
| Winter | 1 | 1.52 | ND | 1.48 | ND |
| 18 | 1.53 | ND | 2.11 | ND | |
| Summer | 1 | 1.50 | 2.25 | 2.38 | 3.57 |
| 18 | 2.04 | 3.06 | 1.34 | 2.01 | |
| 357 | |||||
| Winter | 1 | 1.52 | ND | 0.85 | ND |
| 8 | 0.77 | ND | 0.30 | ND | |
| Summer | 1 | 1.65 | 2.53 | 9.87 | 14.80 |
| 8 | 1.52 | 2.47 | 5.22 | 7.83 | |
| 341 | |||||
| Winter | 1 | 1.36 | ND | 1.69 | ND |
| 16 | 1.45 | ND | 3.04 | ND | |
| Summer | 1 | 1.75 | 2.63 | 5.34 | 8.02 |
| 15 | 1.69 | 2.53 | 0.87 | 1.30 | |
Abbreviations: TB, total bacteria; Cyanos, cyanobacteria; ND, not determined. Rates are given as contacts as well as per cell.
Contact rates between putative cyanomyoviruses (using g20 gene copies as virus particle estimates) and total bacteria in the system were estimated, and specific contact rates were higher in the summer (from 0.66 to 9.87 contacts, with an average of 4.09 contacts cell−1 day−1) than in the winter (0.30 to 3.0 contacts, with an average of 1.74 contacts cell−1 day−1). During the summer, estimates of rates of contact between cyanobacteria and cyanomyoviruses were also very high and ranged from 0.98 to 10.52 (average of 6.13) contacts cell−1 day−1, with the highest rates being found in the surface waters at all stations.
Pearson's correlations of g20 gene copies to biotic and abiotic parameters were determined for both the summer and winter samples (Table 3). Estimated g20 abundance in summer samples was significantly and negatively correlated with depth (−0.77, P = 0.03) and all chlorophyll a size fractions (r = 0.75, P = 0.03, for picophytoplankton; r = 0.87, P = 0.004, for nanophytoplankton; r = 0.77, P = 0.03, for microphytoplankton) while the g20 abundances in winter samples did not significantly correlate with any biotic or abiotic factors.
Table 3.
Pearson correlation coefficients and P values for biotic and abiotic factors affecting putative cyanomyovirus (g20) distribution in Lake Eriea
| Factor | Winter 2009 |
Summer 2009 |
||
|---|---|---|---|---|
| Pearson correlation coefficient | P value | Pearson correlation coefficient | P value | |
| Depth | −0.13 | 0.76 | −0.77 | 0.03 |
| Temp | −0.36 | 0.38 | 0.55 | 0.16 |
| Total bacteria | −0.65 | 0.08 | 0.23 | 0.58 |
| Total Synechococcus | ND | ND | 0.32 | 0.43 |
| Total virus abundance | 0.35 | 0.39 | −0.03 | 0.95 |
| Chl a (0.2-2 μm) | 0.59 | 0.13 | 0.75 | 0.03 |
| Chl a (2-20 μm) | 0.65 | 0.08 | 0.87 | 0.00 |
| Chl a (<20 μm) | 0.68 | 0.06 | 0.77 | 0.03 |
Values considered significant (P<0.05) are in bold. ND, not determined.
DISCUSSION
The goal of this study was to determine if putative cyanomyoviruses were abundant in Lake Erie (using g20 gene abundance as a proxy), and if so, to examine their distribution with respect to biotic and abiotic parameters. The results demonstrate that these viruses are abundant and represent a substantial proportion of the total virus community. They also demonstrate that there is a sufficient population of cyanomyoviruses and their putative hosts to support the interactions and infections necessary to maintain the community. We present these data in the context of the potential ecological and functional roles of cyanomyophage within the lake system.
We used two cruises of opportunity to collect seasonal time point samples—in some cases representing some of the first wintertime microbial estimates for this large lake. Significant seasonal changes were evident in the lake with respect to the abundance of both the bacteria and viruses. The abundance of total virus particles estimated during this study was similar to those found in previous years in Lake Erie: from 3.7 × 107 to 3.7 × 108 ml−1 in 2000 (51) and 2.0 × 107 to 4.1 × 107 ml−1 in 2001 (49). These abundances are comparable to those of other freshwater lakes (50), including the adjacent Lake Ontario (13), although they are higher than virus abundances observed in oligotrophic Lake Superior (39). Seasonal changes in viral abundances are often exhibited in freshwater systems and have been correlated with temperature and bacterial community changes (20). Summer total bacterial abundances were similar to those found in previous years in Lake Erie (8) and to other freshwater systems (39, 42), while summer cyanobacterial counts were higher than those in previous years in Lake Erie (47). During the current study, we were not able to provide data on Synechococcus abundance during winter sampling. However, in February 2007, direct counts by epifluorescence of Synechococcus cells demonstrated (3.7 ± 0.7) × 103 cells ml−1 at station 84 (1 m) and (2.6 ± 0.4) × 103 cells ml−1 in the water column at station 357 (S. W. Wilhelm and G. S. Bullerjahn, unpublished data), suggesting that they remain a seasonally abundant component (∼1 to 10% of the total prokaryotes) within the system.
As introduced previously, there is a substantial literature examining the richness of cyanomyophage in aquatic systems—this includes observations from targeted studies as well as metagenomic assemblages (2, 54). In the current study, high g20 gene densities were found at all stations and in both seasons, with up to 3.1 × 106 g20 genes ml−1 detected in one sample. The highest g20 abundances were found at station 84 in the summer, which may be due to the abundant Synechococcus hosts found in the central basin during summer months (47, 48). These viruses represented 1.8% (0.2 to 4.6%) of the total virus community in summer months and 1.3% (0.4 to 2.3%) of the winter virus community. In the one other study quantitatively targeting g20 for Cyanomyoviridae, cyanomyophage were detected in a Norwegian coastal marine station at up to 7.2 × 103 ml−1, with the lowest abundances during the winter months, but in each case at <0.1% of the total virus population (32). While on the surface these percentages remain low, it must be remembered that virus communities are genetically rich: indeed, ∼90% of the genes within marine virus communities appear novel to science (2). To this end, while numerical dominance does not necessarily indicate ecological relevance, the high abundance of targets suggests not just that these viruses are an active component of the Lake Erie ecosystem but that they may be one of the most abundant virus populations within the microbial community.
A major regulator of the standing stock of viruses is the particle removal rate. In many marine studies, research has shown that virus infectivity and in many cases whole particles are rapidly destroyed under ambient conditions by processes such as exposure to solar radiation (37, 43). As such, in summer months when light penetration through the lake is deep (47), our data point to the need for a constant resupply of these viruses, suggesting high virus production rates. Indeed, if standard decay rates (∼0.5 day−1) and burst sizes (81 cyanophage per lytic infection) are considered proxies (12), our estimates of cyanophage abundance require 2.3 × 103 to 18.8 × 103 Synechococcus cells ml−1 day−1 (∼2% of the Synechococcus population) to be destroyed daily by viruses to maintain virus densities: this is similar to estimates of 1 to 8% within marine surface waters (12). Although this does not seem like a large proportion of the total Synechococcus community, low potential Synechococcus growth rates (with doubling times ranging from 7 h to 7 days [6]) suggest that a significant portion of the carbon flow through picophytoplankton may be diverted by Cyanomyoviridae.
The activity of cyanomyophage in the winter is difficult to estimate as it is likely that their decay rates are lower than in summer months. In the lab, it is well known that aquatic virus samples collected during research cruises remain stable for many years if stored in the dark at 4°C (2). Moreover, ambient solar radiation is significantly reduced in the winter due to the shorter day lengths as well as ice and snow cover. Indeed, in a previous study of an alpine lake, decreases in viral abundance to undetectable levels were attributed to increased UV levels after ice breakage (15). To this end, it appears that maintenance of the high abundance of total viruses, including cyanomyoviruses, observed during the winter months may in part be due to decreased rates of decay. Observations made during sample collection for this study have demonstrated that significant primary production occurs in Lake Erie in both summer and winter months but suggest that winter productivity in this system is driven by diatoms (23; M. R. Twiss, R. M. L. McKay, and H. J. Carrick, personal communication). And, while heterotrophic bacterial activity is high in summer months (8), winter production rates are ∼1% of summer rates (Wilhelm and Bullerjahn, unpublished). How Synechococcus bacteria respond to seasonal changes remains unknown: although abundant here, their reduced numbers in polar environments suggest that they might be dormant during winter months in systems like Lake Erie. If decay rates of viruses are lower in winter months, in sum it suggests a balance between decreased production and loss and allowance for the maintenance of significant standing stock.
The production of virus particles in any environment ultimately depends on the successful interaction between an infectious virus particle and a permissive host cell. It has previously been suggested that at least ∼104 bacterial hosts ml−1 are necessary for the minimum contact rate needed to support viral infection and replication (46). Adequate densities for Synechococcus may be lower than those for other heterotrophic bacteria, however, due to both the lower growth rate for cyanobacteria (40) and the larger size of cells (which increases the rate of virus-host contact). In this study, we found higher virus-host contact rates during the summer than during the winter due to the higher summer densities of both potential hosts and viruses. Cell-specific rates of 77.3 to 152.8 contacts cell−1 day−1 were calculated in the winter and 150.1 to 204.3 contacts cell−1 day−1 in the summer for total bacteria and total viruses in the system, which is higher than what has been previously reported for Lake Erie (4 to 11 contacts cell−1 day−1) (51) and marine systems (0.2 to 4.4 contacts cell−1 day−1) (52) but similar to data for other eutrophic freshwater systems such as Alte Donau (91 to 230 contacts cell−1 day−1) (10) and Lake Plußsee (49 to 180 contacts cell−1 day−1) (42). For putative Cyanomyoviridae, contact rates were ca. 2 orders of magnitude lower than those calculated for the total virus community but were still ≥1 Synechococcus cell−1 day−1. What these contacts represent in terms of real (successful) infections remains to be determined: in only a few studies have researchers attempted to estimate the rate at which contacts result in successful infection (12, 52). It does suggest, however, that for the host cells to persist, there must be a significant diversity of virus-host relationships and likely a continued evolution of resistance to infection within the population (18). The availability of molecular assays to track specific populations should allow for the rate of successful contacts to be examined in future efforts.
As a step toward understanding the distribution of these viruses in this system, we carried out Pearson's correlation analyses on samples from winter and summer months. Although we have a limited sample size in this study, a significant and strong (r = −0.77) negative correlation between g20 densities and depth was observed in the summer samples, consistent with these viruses being produced or localized to surface waters with photosynthetic organisms. This correlation is also consistent with strong correlations to our proxies for phytoplankton biomass: significant and strong correlations were seen between g20 and all size classes of chlorophyll in summer samples. Indeed, the relationship between primary producers and g20 is in part emphasized by the observations that putative Cyanomyoviridae formed the greatest proportion of the virus community at the station with the highest primary production biomass (station 357) during the most productive time of year (summer). That no relationships were seen in winter months is not surprising, as (noted above) we believe that reduced removal rates of particles may play an important role. What is in part surprising about these data is the lack of correlation with specific potential hosts (either Synechococcus or total heterotrophic bacteria) that was observed. However, when one considers that these relationships likely occur across temporal and spatial scales, the survey-style approach to sample collection in this study likely decreases our ability to tease apart these relationships.
One final caveat is that it remains difficult to distinguish between “free” virus signatures in the water column and particle-associated (either on the surface of debris or a cell or in an active infection, lysogen, or prophage) signatures within our data set. Using a filtration-based approach, we attempted to ascertain the proportion of viruses that appear in the cell-associated (unfilterable) group. Our observations suggest that the use of the 1.0-μm filtration did not remove free virus particles from control lysates but that it did reduce the g20 targets in 6 of our 8 field samples that we tested from the winter stations. Since the phage lysate assay suggested no significant difference between the filtered and unfiltered samples, our results suggest that the 1.0-μm filtering process is removing particle-associated (i.e., within cells but also potentially detrital) gene targets. A new hypothesis arising from these observations is that lysogeny may be involved in the maintenance of populations during winter months: this has been a topic of discussion for years, and conflicting evidence suggesting a complex relationship of lysogenic bacteria during winter months has arisen (19, 22, 34, 53).
The requirement for quantitative answers to address ecological questions is an old tenet. In virus ecology, much of the recent efforts have focused on resolving the genetic richness of populations of specific aquatic viruses. Quantification of these viruses is an equally important step toward understanding how they may contribute to the ecology of aquatic environments. In this study, we have demonstrated that a population of well-studied viruses is numerically important within a large lake, laying the groundwork for future efforts to more clearly, and quantitatively, define the interactions between viruses and their hosts.
ACKNOWLEDGMENTS
We thank M. A. Saxton for sample collection as well as the captains and crews of CCGS Griffon and CCGS Limnos for their help and logistical support. We also thank G. S. Bullerjahn, R. M. L. McKay, M. R. Twiss, and the entire WAMBAM research team for comments and assistance.
This research was funded by grants from the National Science Foundation to S.W.W. (OCE-0452409 and OCE-0851113) and a New York Sea Grant (through M. R. Twiss).
Footnotes
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Allender C. J., et al. 2009. Identifying the source of unknown microcystin genes and predicting microcystin variants by linking multiple genes within uncultured cyanobacterial cells. Appl. Environ. Microbiol. 75:3598–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angly F. E., et al. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brittain S. M., et al. 2000. Isolation and characterization of microcystins, cyclic heptapeptide hepatotoxins from a Lake Erie strain of Microcystis aeruginosa. J. Great Lakes Res. 26:241–249 [Google Scholar]
- 4. Brussaard C. P. D., et al. 2008. Global scale processes with a nanoscale drive—from viral genes to oceanic biogeochemical cycles. ISME J. 2:575–578 [DOI] [PubMed] [Google Scholar]
- 5. Buchan A., Hadden M., Suzuki M. T. 2009. Development and application of quantitative-PCR tools for subgroups of the Roseobacter clade. Appl. Environ. Microbiol. 75:7542–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callieri C., Stockner J. G. 2002. Freshwater autotrophic picoplankton: a review. J. Limnol. 61:1–14 [Google Scholar]
- 7. Clasen J. L., Brigden S. M., Payet J. P., Suttle C. A. 2008. Evidence that viral abundance across oceans and lakes is driven by different biological factors. Freshw. Biol. 53:1090–1100 [Google Scholar]
- 8. DeBruyn J. M., Leigh-Bell J. A., McKay R. M. L., Bourbonniere R. A., Wilhelm S. W. 2004. Microbial distributions and the impact of phosphorus on bacterial activity in Lake Erie. J. Great Lakes Res. 30:166–183 [Google Scholar]
- 9. Dorigo U., Jacquet S., Humbert J. F. 2004. Cyanophage diversity, inferred from g20 gene analyses, in the largest natural lake in France, Lake Bourget. Appl. Environ. Microbiol. 70:1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fischer U. R., Velimirov B. 2002. High control of bacterial production by viruses in a eutrophic oxbow lake. Aquat. Microb. Ecol. 27:1–12 [Google Scholar]
- 11. Fuller N. J., Wilson W. H., Joint I. R., Mann N. H. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g20 and its application to PCR-based detection and quantification techniques. Appl. Environ. Microbiol. 64:2051–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garza D. R., Suttle C. A. 1998. The effect of cyanophages on the mortality of Synechococcus spp. and selection for UV resistant viral communities. Microb. Ecol. 36:281–292 [DOI] [PubMed] [Google Scholar]
- 13. Gouvêa S. P., et al. 2006. Phosphorus bioavailability and plankton distributions in Lake Ontario in the aftermath of Hurricane Isabel, September 2003. J. Great Lakes Res. 32:455–470 [Google Scholar]
- 14. Hobbie J. E., Daley R. J., Jasper S. 1977. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hofer J., Sommaruga R. 2001. Seasonal dynamics of viruses in an alpine lake: importance of filamentous forms. Aquat. Microb. Ecol. 26:1–11 [Google Scholar]
- 16. Ivanikova N. V., et al. 2008. Picoplanktonic cyanobacteria in Lakes Superior and Erie: phylogenies of endemic populations and cultured isolates. Verhandlungen Internationale Vereinigung fur theoretische und angewandte Limnologie 30:459–465 [Google Scholar]
- 17. Kibbe W. A. 2007. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35:W43–W46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lennon J. T., Martiny J. B. H. 2008. Rapid evolution buffers ecosystem impacts of viruses in a microbial food web. Ecol. Lett. doi:10.1111/j.1461–0248.2008.01225.x [DOI] [PubMed] [Google Scholar]
- 19. Lisle J. T., Priscu J. C. 2004. The occurrence of lysogenic bacteria and microbial aggregates in the lakes of the McMurdo Dry Valleys. Microb. Ecol. 47:427–439 [DOI] [PubMed] [Google Scholar]
- 20. Lymer D., et al. 2008. Temporal variation in freshwater viral and bacterial community composition. Freshw. Biol. 53:1163–1175 [Google Scholar]
- 21. Marston M. F., Sallee J. L. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl. Environ. Microbiol. 69:4639–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McDaniel L., Houchin L. A., Williamson S. J., Paul J. H. 2002. Plankton blooms: lysogeny in marine Synechococcus. Nature 415:496. [DOI] [PubMed] [Google Scholar]
- 23. McKay R. M. L., Beall B. F. N., Bullerjahn G. S., Woityra W. C. 2011. Winter limnology on the Great Lakes: the role of the U.S. Coast Guard. J. Great Lakes Res. 37:207–210 [Google Scholar]
- 24. Morrison T. B., Weis J. J., Wittwer C. T. 1998. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 24:954–962 [PubMed] [Google Scholar]
- 25. Muhling M., et al. 2005. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ. Microbiol. 7:499–508 [DOI] [PubMed] [Google Scholar]
- 26. Munawar M., Edsall T., Munawar I. F. 1999. State of Lake Erie: past, present, and future. Backhuys Publishers, Leiden, Netherlands [Google Scholar]
- 27. Murray A. G., Jackson G. A. 1992. Viral dynamics: a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Prog. Ser. 89:103–116 [Google Scholar]
- 27a. Noble R. T., Fuhrman J. A. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113–118 [Google Scholar]
- 28. Rinta-Kanto J. M., et al. 2009. Lake Erie Microcystis: relationship between microcystin production, dynamics of genotypes and environmental parameters in a large lake. Harmful Algae 8:665–673 [Google Scholar]
- 29. Rinta-Kanto J. M., et al. 2005. Quantification of toxic Microcystis spp. during the 2003 and 2004 blooms in Western Lake Erie. Environ. Sci. Technol. 39:4198–4205 [DOI] [PubMed] [Google Scholar]
- 30. Rinta-Kanto J. M., et al. 2009. The diversity and distribution of toxigenic Microcystis spp. in present day and archived pelagic and sediment samples from Lake Erie. Harmful Algae 8:385–394 [Google Scholar]
- 31. Rinta-Kanto J. M., Wilhelm S. W. 2006. Diversity of microcystin-producing cyanobacteria in spatially isolated regions of Lake Erie. Appl. Environ. Microbiol. 72:5083–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandaa R.-A., Larsen A. 2006. Seasonal variations in virus-host populations in Norwegian coastal waters: focusing on the cyanophage community infecting marine Synechococcus spp. Appl. Environ. Microbiol. 72:4610–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Short C. M., Suttle C. A. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl. Environ. Microbiol. 71:480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stopar D., Cerne A., Zeigman M., Ploljsak-Prijatelj M., Turk V. 2004. Viral abundance and high proportion of lysogens suggest that viruses are important members of the microbial community in the Gulf of Trieste. Microb. Ecol. 47:1–8 [DOI] [PubMed] [Google Scholar]
- 35. Sullivan M. B., et al. 2008. Portal protein diversity and phage ecology. Environ. Microbiol. 10:2810–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suttle C. A. 2000. Cyanophages and their role in the ecology of cyanobacteria, p. 563–589. In Whitton B. A., Potts M. (ed.), The ecology of cyanobacteria: their diversity in time and space. Kluwer, Boston, MA [Google Scholar]
- 37. Suttle C. A., Chen F. 1992. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58:3721–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takashima Y., et al. 2007. Development and application of quantitative detection of cyanophages phylogenetically related to cyanophage Ma-LMM01 infecting Microcystis aeruginosa in fresh water. Microb. Environ. 22:207–213 [Google Scholar]
- 39. Tapper M. A., Hicks R. E. 1998. Temperate viruses and lysogeny in Lake Superior bacterioplankton. Limnol. Oceanogr. 43:95–103 [Google Scholar]
- 40. Waterbury J. B., Valois F. W. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weinbauer M. G., Chen F., Wilhelm S. W. 2011. Virus-mediated redistribution and partitioning of carbon in the global oceans, p. 54–56. In Jiao N., Azam F., Sanders S. (ed.), Microbial carbon pump in the ocean. Science/AAAS, Washington, DC [Google Scholar]
- 42. Weinbauer M. G., Höfle M. G. 1998. Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic lake. Appl. Environ. Microbiol. 64:431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weinbauer M. G., Wilhelm S. W., Suttle C. A., Pledger R. J., Mitchell D. L. 1999. Sunlight-induced DNA damage and resistance in natural viral communities. Aquat. Microb. Ecol. 17:111–120 [Google Scholar]
- 44. Welschmeyer N. A. 1994. Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol. Oceanogr. 39:1985–1992 [Google Scholar]
- 45. Wetzel R. G. 2001. Limnology. Academic Press, New York, NY [Google Scholar]
- 46. Wiggins B. A., Alexander M. 1985. Minimum bacterial density for bacteriophage replication: implications for significance of bacteriophages in natural ecosystems. Appl. Environ. Microbiol. 49:19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilhelm S. W., et al. 2006. Seasonal hypoxia and the genetic diversity of prokaryote populations in the central basin hypolimnion of Lake Erie: evidence for abundant cyanobacteria and photosynthesis. J. Great Lakes Res. 32:657–671 [Google Scholar]
- 48. Wilhelm S. W., et al. 2006. Marine and freshwater cyanophages in a Laurentian Great Lake: evidence from infectivity assays and molecular analyses of g20 genes. Appl. Environ. Microbiol. 72:4957–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilhelm S. W., et al. 2003. The effect of phosphorus amendments on present day plankton communities in pelagic Lake Erie. Aquat. Microb. Ecol. 32:275–285 [Google Scholar]
- 50. Wilhelm S. W., Matteson A. R. 2008. Freshwater and marine virioplankton: a brief overview of commonalities and differences. Freshw. Biol. 53:1076–1089 [Google Scholar]
- 51. Wilhelm S. W., Smith R. E. H. 2000. Bacterial carbon production in Lake Erie is influenced by viruses and solar radiation. Can. J. Fish. Aquat. Sci. 57:317–326 [Google Scholar]
- 52. Wilhelm S. W., Weinbauer M. G., Suttle C. A., Jeffrey W. H. 1998. The role of sunlight in the removal and repair of viruses in the sea. Limnol. Oceanogr. 43:586–592 [Google Scholar]
- 53. Williamson S. J., Houchin L. A., McDaniel L., Paul J. H. 2002. Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microbiol. 68:4307–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Williamson S. J., et al. 2008. The Sorcerer II global ocean sampling expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3:e1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhong Y., Chen F., Wilhelm S. W., Poorvin L., Hodson R. E. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68:1576–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]