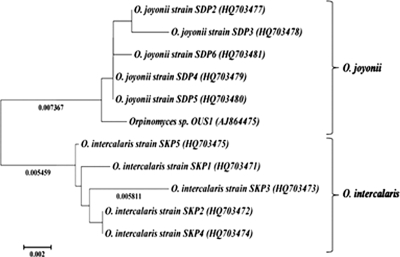
Fig. 5.
Neighbor-joining phylogenetic tree of Orpinomyces spp. obtained from the sequences of the LSU. The evolutionary history was inferred using the neighbor-joining method (19). The optimal tree with the sum of branch length of 0.03107997 is shown. The tree is drawn to scale, with branch lengths (below the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (21) and are in the units of the number of base substitutions per site. The analysis involved 11 nucleotide sequences. The codon positions included were the 1st, 2nd, 3rd, and noncoding positions. All positions containing gaps and missing data were eliminated. There were a total of 743 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (22).