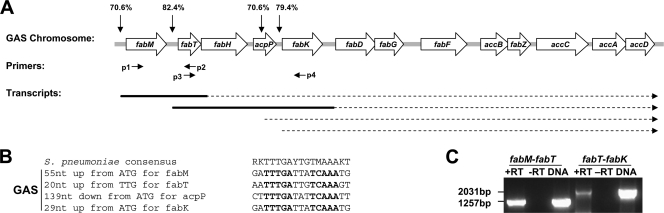
Fig. 3.
Fatty acid biosynthesis (fab) region in GAS. (A) Predicted transcriptional organization for the fab region in GAS. Vertical arrows point to the predicted binding sites for the transcriptional repressor FabT in MGAS5005 with % homology to the consensus S. pneumoniae binding site shown above each arrow. Small horizontal arrows indicate location of primers used for detection of fab transcripts in GAS (p1 to p4). Dashed lines depict predicted transcripts for the fab region, and bold lines delineate the transcripts detected by RT-PCR (see panel C). (B) Sequence and location of the predicted FabT binding sites in MGAS5005. The palindromic sequence repeat that is conserved in GAS is delineated in bold letters. (C) RT-PCR analysis of transcripts for the fab region in GAS. The reaction was performed with (+RT) or without (−RT) reverse transcriptase using MGAS2221 RNA and primers p1 and p2 to identify a transcript that includes both fabM and fabT (fabM-fabT) or primers p3 and p4 to identify a transcript including fabT, fabH, acpP and fabK (fabT-fabK). DNA of MGAS2221 was used as a positive control for the PCR.