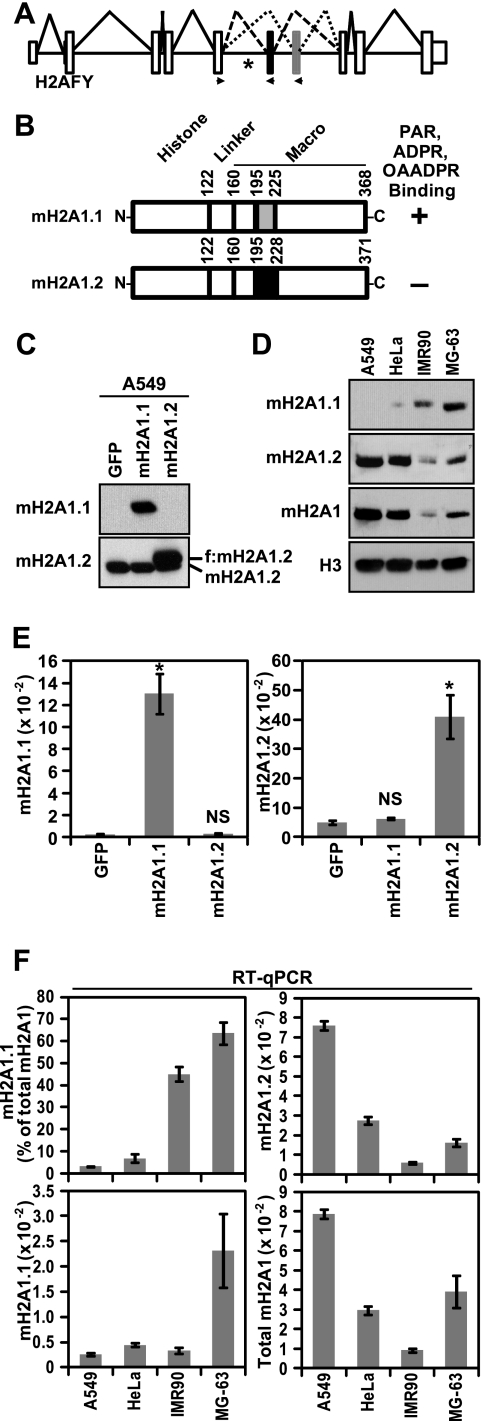
Fig. 1.
Differences in macroH2A1 alternative splicing and protein levels across four cell lines. (A) Diagram depicting the structure and splicing of the gene encoding macroH2A1, H2AFY. The small white boxes, the large white boxes and the solid horizontal lines represent untranslated regions, coding exons and introns, respectively. The macroH2A1.2- and macroH2A1.1-specific exons are black and gray, respectively. Solid, dashed, and dotted lines depict constitutive and macroH2A1.2 or macroH2A1.1 alternative splicing events, respectively. Locations of expression primers used for splice variant-specific qPCR are indicated with arrowheads. The location of a QKI binding site is indicated by an asterisk (27). (B) Schematic depicting the two alternatively spliced macroH2A1 variants. The locations of amino acids encoded by the variable exons are indicated by gray and black boxes for macroH2A1.1 (mH2A1.1) and macroH2A1.2 (mH2A1.2), respectively. Amino acid positions marking the boundaries of the histone-like domain, the linker region, the variable exons, and the macro domain are indicated. The plus and minus signs indicate the affinity or lack of affinity for ADPR-based ligands, respectively. OAADPR, O-acetyl-ADPR. (C) The specificity of anti-macroH2A1.1 and anti-macroH2A1.2 antibodies was determined by immunoblots of A549 cells expressing GFP, Flag-tagged macroH2A1.1, or Flag-tagged macroH2A1.2 (f:mH2A1.2). The location of endogenous and ectopic macroH2A1 is indicated. (D) Immunoblots demonstrating the differences in macroH2A1.1, macroH2A1.2, and total macroH2A protein levels in four cell lines. Anti-histone H3 was used as a loading control. (E) Histograms depicting the specificity of the macroH2A1.1 and macroH2A1.2 primer sets in the RT-qPCR assay with A549 cells expressing the indicated factors as described for panel C. Error bars represent standard errors of the means (SEMs) for four biological replicates. P values are the result of a two-tailed Student's t test. *, P < 0.001; NS, not significant. (F) Histograms depicting the macroH2A1.1 and macroH2A1.2 levels across four cell lines. The data are expressed as either ACTB normalized (mH2A1.1 and mH2A1.2), the sum of macroH2A1.1 and macroH2A1.2 (Total mH2A1), or the percentage of total macroH2A1 that has been spliced as macroH2A1.1 [mH2A1.1 (% of total mH2A)] as indicated. Error bars represent the SEMs for three biological replicates.