Abstract
We present a novel denaturing gradient gel electrophoresis (DGGE) method which characterizes multiclonal communities of Staphylococcus aureus. The spa PCR-based DGGE method simultaneously separates strains that differ in only one base, thereby revealing multiclonal colonization and infections.
TEXT
Staphylococcus aureus causes a wide range of infections and is responsible for a considerable portion of hospital-acquired infections.
In 30% to 70% of healthy individuals, S. aureus is a transient or persisting part of the residential flora (5, 10).
Cespedes et al. investigated the frequency of simultaneous nasal carriage of multiple S. aureus strains by picking three bacterial colonies from plates derived from each colonized individual. Fewer than 7% of them were predicted to carry >1 strain (1). The simultaneous presence of an invasive and a carrier strain of methicillin-resistant S. aureus (MRSA) in one individual was reported by Soderquist and Berglund (12). The issue of multiclonal colonization is important, but conventional laboratory methods for detection of S. aureus are based on culture of a single colony. This might result in the identification of an antibiotic-susceptible commensal strain rather than a second, more resistant strain, which may impair the antibiotic treatment and bias epidemiological conclusions.
We have developed a species-specific denaturing gradient gel electrophoresis (DGGE) method for S. aureus, utilizing spa, to characterize multiclonal colonization and infection. The novel assay was used to investigate a MRSA outbreak, revealing colonization with two different strains in some of the individuals.
spa typing has been documented to be a useful tool in investigations of MRSA epidemiology (7) and for studies of S. aureus transmission (6). Thus, we based our DGGE method on spa and primer pairs, described by Kahl et al. (4), which were modified for DGGE analyses by the attachment of a GC clamp (11) at either the forward or the reverse primer. Annealing temperature and MgCl2 concentrations for two primer combinations were optimized for PCR specificity and efficiency (data not shown).
Eight S. aureus isolates of known spa types were acquired from the Microbiology Laboratory, Ryhov County Hospital, Jönköping, Sweden. All isolates contained 10 spa repeats (Table 1). The isolates were suspended in 200 μl PCR-grade water (Sigma-Aldrich, St. Louis, MO) and lysed for 10 min at 95°C. DNA was purified using the MagAttract DNA Mini M48 kit on BioRobot M48 (Qiagen, Hilden, Germany).
Table 1.
spa types and repeat succession of S. aureus strains used in the evaluation of the spa-DGGE method
| spa type | Repeat succession |
|---|---|
| t015 | 08-16-02-16-34-13a-17-34-16-34 |
| t050 | 08-16-02-16-34-34a-17-34-16-34 |
| t008 | 11-19-12-21a-17-34-24-34-22-25 |
| t064 | 11-19-12-05a-17-34-24-34-22-25 |
| t002 | 26-23-17-34-17-20-17-12-17-16 |
| t012 | 15-12-16-02-16-02-25-17-24-24 |
| t355 | 07-56-12-17-16-16-33-31-57-12 |
| t3061 | 07-21-17-34-13-34-34-13-33-13 |
Repeat numbers 13 and 34 as well as 5 and 21 differ in only one base, respectively.
The most stringent separation of the PCR products (not shown) was achieved when the GC clamp was attached to the forward primer spa-1113f-GC. The optimized PCR mixture contained 12.5 μl HotStar Mastermix (Qiagen), 1.5 mM MgCl2 (Roche, Mannheim, Germany), 0.2 nmol forward primer spa-1113f-GC (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGTAAAGACGATCCTTCGGTGAGC-3′; GC clamp indicated in bold), and 0.2 nmol reverse primer spa-1514r (5′-CAGCAGTAGTGCCGTTTGCTT-3′) (TIB Molbiol, Berlin, Germany). The reaction conditions were 15 min at 95°C, followed by 45 cycles of 30 s at 94°C, 30 s at 61°C, and 60 s at 72°C, with a final extension for 10 min at 72°C.
The PCR products were analyzed using a DCode universal mutation detection system (Bio-Rad Laboratories Inc., Hercules, CA). Polyacrylamide gradient gels (160 by 160 by 1 mm) composed of 37.5:1 acrylamide-bisacrylamide (7%) and 1× TAE (40 mM Tris-HCl, 20 mM sodium acetate, 1 mM EDTA [pH 8.3]) with 15% to 50% denaturant were cast with the aid of a gradient former (Bio-Rad Laboratories Inc.). The gels were polymerized with 10 μl of TEMED (N,N,N′,N′-tetramethylethylenediamine) and 173 μl of 10% ammonium persulfate. Two hundred forty nanograms of DNA was loaded into each well, and the gels were run in 1× TAE buffer at 62°C for 14 h at 130 V. Gels were stained in 1× TAE buffer containing SYBRGold (Invitrogen, Paisley, United Kingdom) for 40 min and visualized in UV transillumination using a charge-coupled device (CCD) camera (LAS-3000; FujiFilm, Tokyo, Japan).
The detection limit was 20 gene copies of spa per reaction (not shown), and the method could discriminate PCR products of the same length from a mixture of eight strains (Fig. 1). In the developed spa-DGGE assay, it was possible to simultaneously amplify DNA of two different spa types (t064 and t355, respectively) in a mixture with a concentration difference of 1:1,000 (not shown). By using conventional cultivation methods, an extreme number of colonies would be required to achieve a comparable sensitivity to detect multiclonality.
Fig. 1.
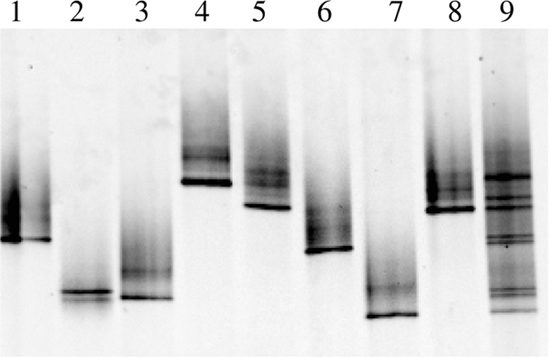
spa-DGGE analysis of the eight different strains (lanes 1 to 8, t064, t355, t012, t3061, t050, t008, t002, and t015, respectively; lane 9, a mixture of equal amounts of eight PCR products).
An outbreak of MRSA occurred at a nursing home in 2009. Screening for possible colonization of patients (n = 24), staff (n = 50), and family members of colonized staff (n = 6) was performed. Swab (Copan, Brescia, Italy) samples (n = 229) from throat, anterior nares, and groin were cultured in broth, and the presence of MRSA was verified in 37 broth samples from 12 individuals, by detection of nuc and mecA according to the method of Nilsson et al. (9). One MRSA isolate from each individual was spa typed as described previously (3, 4). Two different but closely related spa types were isolated from the 12 individuals (4 patients, 4 staff, and 4 relatives of staff) of the outbreak. Nine individuals were colonized with S. aureus of spa type t015, and three were colonized with t069 isolates.
One milliliter of broth samples was centrifuged (10,000 × g, 3 min), and DNA extraction was performed as described above. When 28 available broth samples positive by culture for MRSA were analyzed by spa-DGGE, all samples were confirmed to contain either spa type t015 or spa type t069. Besides, in broths from three individuals, spa types t015 and t069 were detected simultaneously (Fig. 2). Dual colonization of MRSA was indicated in samples from the groin and the throat in one of these individuals and in the throat of a second. The remaining samples from these two individuals contained only spa type t069. In the third individual, the samples from throat and nares contained t069 and t015, respectively.
Fig. 2.
Broth samples from the outbreak containing MRSA isolates of spa type t015 or t069 were analyzed by the spa-DGGE method. In samples from two individuals, both spa types, t015 and t069, were detected (lanes 2 and 3). In lanes 4 to 7, the samples contain spa type t015, and in lanes 8 to10, the samples contain spa type t069. In lane 1, PCR products from both spa types, t015 and t069, are mixed and analyzed as a control.
S. aureus multiclonality is rarely studied, although multiclonal infections occur and such infections may indeed affect the selection of antimicrobial therapy and might impede the treatment of serious infections (2). Cespedes et al. showed by culture of three colonies from each sample that fewer than 7% of the population was colonized by more than one strain in the anterior nares (1). However, their approach will probably reveal only major clones. Mongkolrattanothai et al. found two genetically distinct S. aureus strains in 25% of nasal and perianal swab samples from children when 4 to 15 colonies were picked from each culture (8). Thus, there is, using culture, still a risk of underestimating the diversity of S. aureus.
To conclude, we describe a sensitive molecular method with high discriminatory power useful in clinical samples for multiclonal characterization of S. aureus colonization and infection. The method offers the potential to become a valuable epidemiological tool as well as a tool for the investigation of S. aureus infections.
Acknowledgments
We acknowledge Andrea Johansson and Sofia Lundin for their technical assistance.
This work was in part supported by the Swedish Society of Medicine, Futurum, and the Research Council of South-East Sweden (FORSS).
Footnotes
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Cespedes C., et al. 2005. The clonality of Staphylococcus aureus nasal carriage. J. Infect. Dis. 191:444–452 [DOI] [PubMed] [Google Scholar]
- 2. Goerke C., et al. 2007. High phenotypic diversity in infecting but not in colonizing Staphylococcus aureus populations. Environ. Microbiol. 9:3134–3142 [DOI] [PubMed] [Google Scholar]
- 3. Harmsen D., et al. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kahl B. C., Mellmann A., Deiwick S., Peters G., Harmsen D. 2005. Variation of the polymorphic region X of the protein A gene during persistent airway infection of cystic fibrosis patients reflects two independent mechanisms of genetic change in Staphylococcus aureus. J. Clin. Microbiol. 43:502–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kluytmans J., van Belkum A., Verbrugh H. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matussek A., Taipalensuu J., Einemo I. M., Tiefenthal M., Lofgren S. 2007. Transmission of Staphylococcus aureus from maternity unit staff members to newborns disclosed through spa typing. Am. J. Infect. Control 35:122–125 [DOI] [PubMed] [Google Scholar]
- 7. Melin S., et al. 2009. Epidemiological typing of methicillin-resistant Staphylococcus aureus (MRSA): spa typing versus pulsed-field gel electrophoresis. Scand. J. Infect. Dis. 41:433–439 [DOI] [PubMed] [Google Scholar]
- 8. Mongkolrattanothai K., et al. 2011. Simultaneous carriage of multiple genotypes of Staphylococcus aureus in children. J. Med. Microbiol. 60:317–322 [DOI] [PubMed] [Google Scholar]
- 9. Nilsson P., Alexandersson H., Ripa T. 2005. Use of broth enrichment and real-time PCR to exclude the presence of methicillin-resistant Staphylococcus aureus in clinical samples: a sensitive screening approach. Clin. Microbiol. Infect. 11:1027–1034 [DOI] [PubMed] [Google Scholar]
- 10. Nilsson P., Ripa T. 2006. Staphylococcus aureus throat colonization is more frequent than colonization in the anterior nares. J. Clin. Microbiol. 44:3334–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. 1989. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. U. S. A. 86:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soderquist B., Berglund C. 2008. Simultaneous presence of an invasive and a carrier strain of methicillin-resistant Staphylococcus aureus (MRSA) in a family. Scand. J. Infect. Dis. 40:987–989 [DOI] [PubMed] [Google Scholar]



