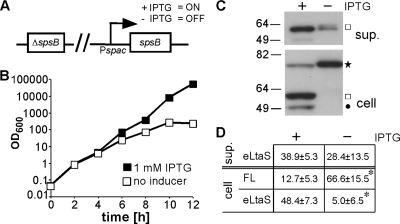
Fig. 2.
S. aureus signal peptidase SpsB is required for efficient LtaS processing. (A) Schematic representation of S. aureus strain ANG2009, with IPTG-inducible spsB expression. (B) Bacterial growth curves. Washed overnight cultures of S. aureus strain ANG2009, with inducible spsB expression, were back diluted into medium with or without IPTG, and bacterial growth was monitored by determining OD600 readings at the indicated time points. Four hours after the initial dilution, cells were washed and back diluted in fresh medium with or without IPTG, and at the 8-h time point, cultures were diluted a second time to maintain them in the logarithmic growth phase. (C) LtaS detection by Western blotting. At the 8-h time point, supernatant (sup.) and whole-cell (cell) protein samples were prepared, and the LtaS protein was detected by Western blotting using the LtaS-specific antibody. A star indicates the full-length LtaS protein, a square the cleaved eLtaS protein, and a filled circle the unspecific Sbi protein band. Note that the Sbi protein itself is cleaved after its transport across the membrane by the signal peptidase SpsB, and hence the absence of the Sbi band in the spsB depletion strain might be due to its instability if it is not properly processed. (D) Densitometry analysis. Densitometry analysis was performed as described in the legend to Fig. 1D, and the average values and standard deviations for seven experiments are shown. Two-tailed two-value equal-variance Student's t test was performed, and statistically significant differences (P < 0.01) are indicated with an asterisk.