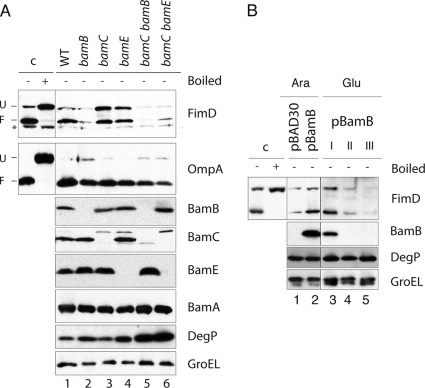
Fig. 4.
FimD folding in E. coli null mutants of nonessential lipoproteins of the BAM complex. (A) Western blot analysis of whole-cell protein extracts from the E. coli UT5601 (WT) strain and isogenic ΔbamB, ΔbamC, ΔbamE, ΔbamB ΔbamC, and ΔbamC ΔbamE null mutants grown statically in BHI medium. Western blots were developed with anti-FimD, anti-OmpA, anti-BamB, anti-BamC, anti-BamE, anti-BamA, anti-DegP, and anti-GroEL antibodies, as indicated on the right of each panel. The mobilities of the protein bands corresponding to folded (F) and unfolded (U) FimD and OmpA are labeled on the left. A nonspecific cross-reactive band against anti-FimD serum is also indicated with an asterisk. Samples loaded in lanes 1 to 6 were not boiled except for detection of BamA. (B) Complementation of the E. coli ΔbamB null mutant with plasmid pBamB (bearing bamB under control of the PBAD promoter). Western blot analysis of whole-cell protein extracts from ΔbamB null mutant bacteria carrying plasmid vector pBAD30 (lane 1) or pBamB (lanes 2 to 5) developed with anti-FimD, anti-BamB, anti-DegP, and anti-GroEL antibodies. Bacteria were cultured in medium with l-arabinose (Ara) (lanes 1 and 2) to induce the PBAD promoter or with d-glucose (Glu) (lanes 3 to 5) to deplete BamB bacteria previously grown with l-arabinose. For depletion in the induced E. coli ΔbamB (pBamB) strain, bacteria were grown with d-glucose and maintained in exponential growth for ∼8 h by repeated dilutions in fresh medium when the culture reached an OD600 of ∼0.5 (indicated as I, II, and III). In both panels, control samples (c) of FimD were obtained from a culture of the WT strain.